Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
X-Ray Structure Of Sars-Cov-2 Main Protease Covalently Bound To Inhibitor Grl-050-22 At 1.16 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: A1C3L |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease Covalently Bound To Inhibitor Grl-062-22 At 1.65 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: A1C3M, NA |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease V186G Covalently Bound To Compound Grl-051-22 At 1.3 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1BFE |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease Covalently Bound To Compound Grl-051-22 At 1.75 A.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1BFE |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease T190I Covalently Bound To Compound Grl-051-22 At 1.5 A
Organism: Severe acute respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1BFE, NA |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease Covalently Bound To Compound Grl-050-23 At 1.6 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN Ligands: DMS, A1BMU, NA |
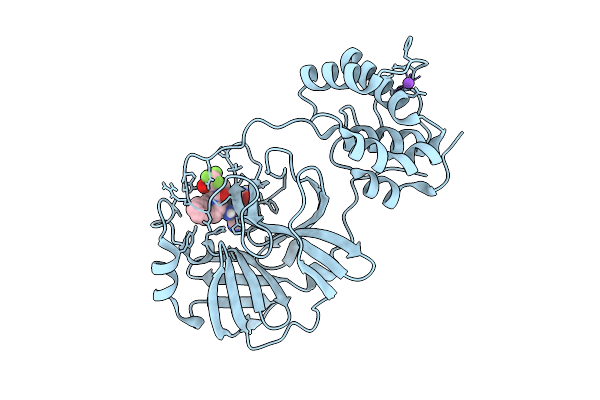 |
X-Ray Structure Of Sars-Cov-2 Main Protease M49I Covalently Bound To Inhibitor Grl-051-22 At 1.50 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN Ligands: A1BFE, NA |
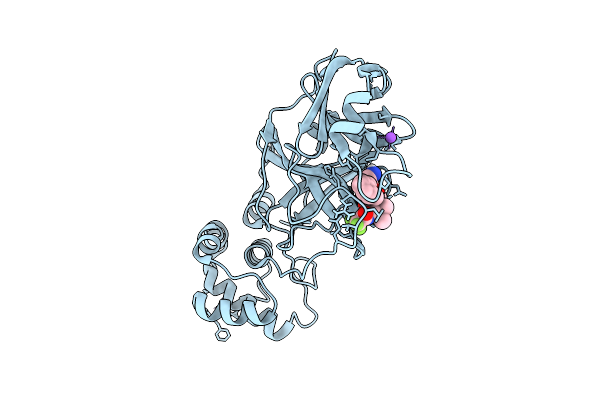 |
X-Ray Structure Of Sars-Cov-2 Main Protease M165I Covalently Bound To Inhibitor Grl-051-22 At 1.90 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN Ligands: A1BFE, NA |
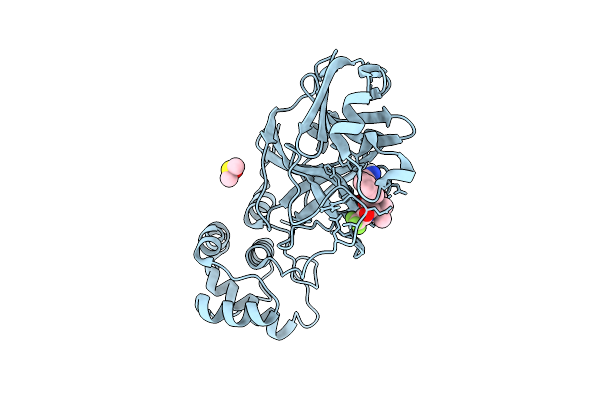 |
X-Ray Structure Of Sars-Cov-2 Main Protease V186F Covalently Bound To Inhibitor Grl-051-22 At 1.60 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1BFE, DMS |
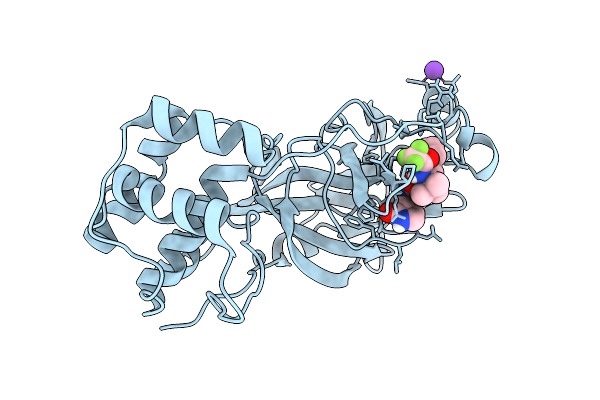 |
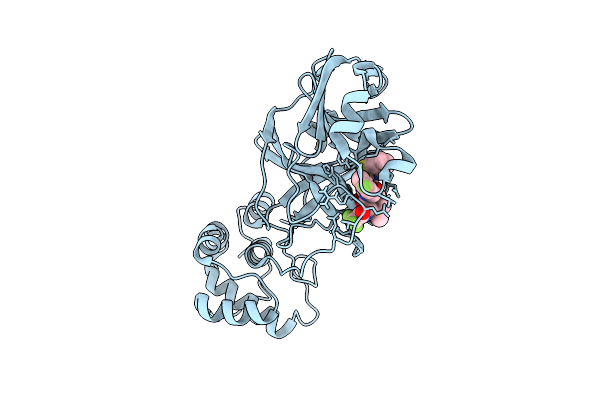
X-Ray Structure Of Sars-Cov-2 Main Protease V186I Covalently Bound To Inhibitor Grl-051-22 At 1.90 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1BFE, K |
 |
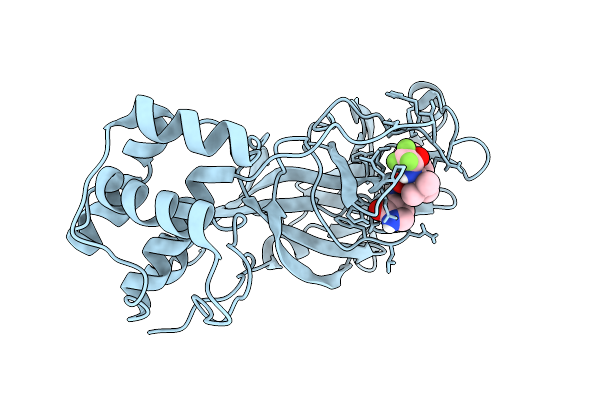
X-Ray Structure Of Sars-Cov-2 Main Protease V186F Covalently Bound To Inhibitor Grl-050-23 At 1.55 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1BMU |
 |
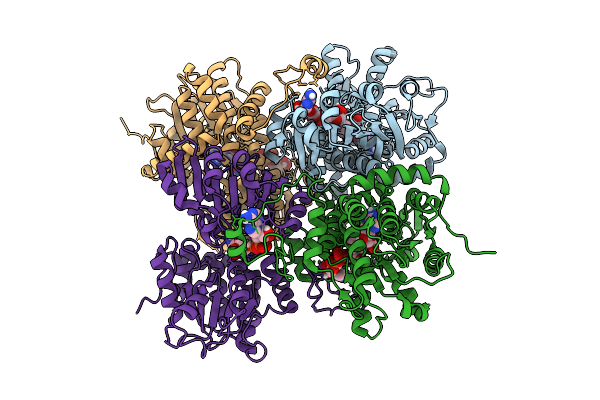
X-Ray Structure Of Sars-Cov-2 Main Protease V186G Covalently Bound To Inhibitor Nirmatrelvir At 1.81 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: 4WI |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: HYDROLASE Ligands: NAD, ADN |
 |
X-Ray Structure Of Human Ship1 Ptase-C2 Domains Covalently Bound To Treat-Ad (Tad) Compound Tad-58547
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2024-10-23 Classification: LIPID BINDING PROTEIN Ligands: X3F, DMS |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease Covalently Bound To Inhibitor Grl-190-21 At 1.90 A.
Organism: Severe acute respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-10-23 Classification: VIRAL PROTEIN Ligands: X4N |
 |
Targeting Enterococcus Faecalis Hmg-Coa Reductase With A Novel Non-Statin Inhibitor
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-09-28 Classification: ANTIMICROBIAL PROTEIN Ligands: SO4, MES, PEG, CA, EDO |
 |
Targeting Enterococcus Faecalis Hmg-Coa Reductase With A Novel Non-Statin Inhibitor
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2022-09-21 Classification: ANTIBIOTIC Ligands: NAP, GOL, CA, ACT, HMG, MEV |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease Covalently Modified By Compound Grl-017-20
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-09-29 Classification: VIRAL PROTEIN Ligands: 4IJ |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease Covalently Modified By Compound Grl-091-20
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-09-29 Classification: VIRAL PROTEIN Ligands: 4I9, NA |
 |
X-Ray Structure Of Sars-Cov Main Protease Covalently Modified By Compound Grl-0686
Organism: Severe acute respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2021-09-29 Classification: VIRAL PROTEIN Ligands: 4IO, DMS |