Search Count: 141
 |
X-Ray Structure Of Sars-Cov-2 Main Protease V186G Covalently Bound To Compound Grl-051-22 At 1.3 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1BFE |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease Covalently Bound To Compound Grl-051-22 At 1.75 A.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1BFE |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease T190I Covalently Bound To Compound Grl-051-22 At 1.5 A
Organism: Severe acute respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1BFE, NA |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease Covalently Bound To Compound Grl-050-23 At 1.6 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN Ligands: DMS, A1BMU, NA |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease M49I Covalently Bound To Inhibitor Grl-051-22 At 1.50 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN Ligands: A1BFE, NA |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease M165I Covalently Bound To Inhibitor Grl-051-22 At 1.90 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN Ligands: A1BFE, NA |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease V186F Covalently Bound To Inhibitor Grl-051-22 At 1.60 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1BFE, DMS |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease V186I Covalently Bound To Inhibitor Grl-051-22 At 1.90 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1BFE, K |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease V186F Covalently Bound To Inhibitor Grl-050-23 At 1.55 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1BMU |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease V186G Covalently Bound To Inhibitor Nirmatrelvir At 1.81 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: 4WI |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: HYDROLASE Ligands: NAD, ADN |
 |
X-Ray Structure Of Human Ship1 Ptase-C2 Domains Covalently Bound To Treat-Ad (Tad) Compound Tad-58547
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2024-10-23 Classification: LIPID BINDING PROTEIN Ligands: X3F, DMS |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease Covalently Bound To Inhibitor Grl-190-21 At 1.90 A.
Organism: Severe acute respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-10-23 Classification: VIRAL PROTEIN Ligands: X4N |
 |
Organism: Pseudomonas sp. 'mevalonii'
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-03-20 Classification: STRUCTURAL PROTEIN Ligands: COA, MEV, SO4, NAD |
 |
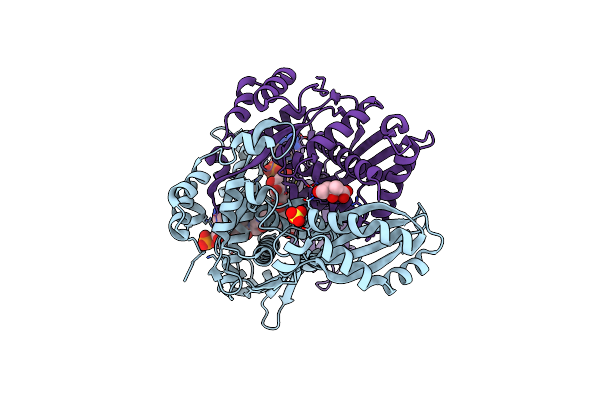
Organism: Pseudomonas sp. 'mevalonii'
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-03-20 Classification: OXIDOREDUCTASE Ligands: ZKE, SO4, XII |
 |
Structure Of Pmhmgr Bound To Mevalonate, Coa And Nad 5 Minutes After Reaction Initiation At Ph 9
Organism: Pseudomonas sp. 'mevalonii'
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2024-03-20 Classification: OXIDOREDUCTASE Ligands: ZKE, COA, A1AFY, SO4, NAD, NAI, MEV |
 |
Targeting Enterococcus Faecalis Hmg-Coa Reductase With A Novel Non-Statin Inhibitor
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-09-28 Classification: ANTIMICROBIAL PROTEIN Ligands: SO4, MES, PEG, CA, EDO |
 |
Targeting Enterococcus Faecalis Hmg-Coa Reductase With A Novel Non-Statin Inhibitor
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2022-09-21 Classification: ANTIBIOTIC Ligands: NAP, GOL, CA, ACT, HMG, MEV |
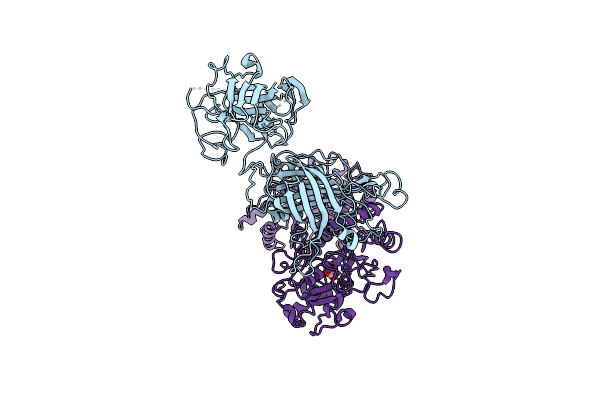 |
The Cryoem Structure Of Lbpb From N. Gonorrhoeae In Complex With Lactoferrin
Organism: Neisseria gonorrhoeae, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-12-01 Classification: MEMBRANE PROTEIN/TRANSPORT PROTEIN Ligands: FE, BCT |
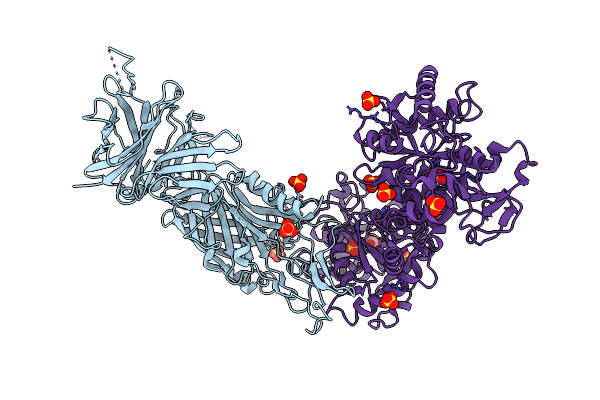 |
The Crystal Structure Of Lactoferrin Binding Protein B (Lbpb) From Neisseria Meningitidis In Complex With Human Lactoferrin
Organism: Neisseria meningitidis serogroup b, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2021-10-13 Classification: METAL TRANSPORT Ligands: SO4, FE, BCT |

