Search Count: 18
 |
Organism: Pseudomonas aeruginosa pao1
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: RIBOSOME Ligands: ZN, MG |
 |
Conformational Landscape Of The Type V-K Crispr-Associated Transposonintegration Assembly Cast V-K Composite Map
Organism: Scytonema hofmannii
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: DNA BINDING PROTEIN Ligands: MG, ZN, ATP |
 |
Conformational Landscape Of The Type V-K Crispr-Associated Transposonintegration Assembly Cast V-K Cas12K Domain Local-Refinement Map
Organism: Scytonema hofmannii
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: DNA BINDING PROTEIN Ligands: MG, ZN |
 |
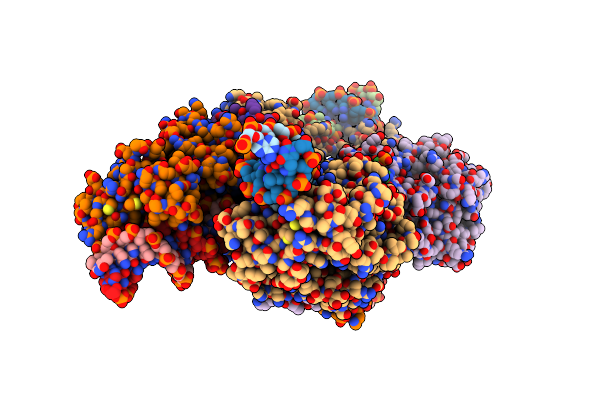
Conformational Landscape Of The Type V-K Crispr-Associated Transposonintegration Assembly Cast V-K Tnsc Domain Local-Refinement Map
Organism: Scytonema hofmannii
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: DNA BINDING PROTEIN Ligands: MG, ATP |
 |
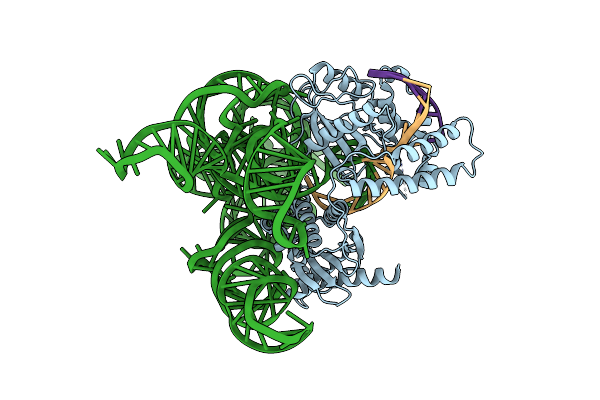
Conformational Landscape Of The Type V-K Crispr-Associated Transposonintegration Assembly Cast V-K Tnsb Domain Local-Refinement Map
Organism: Scytonema hofmannii
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
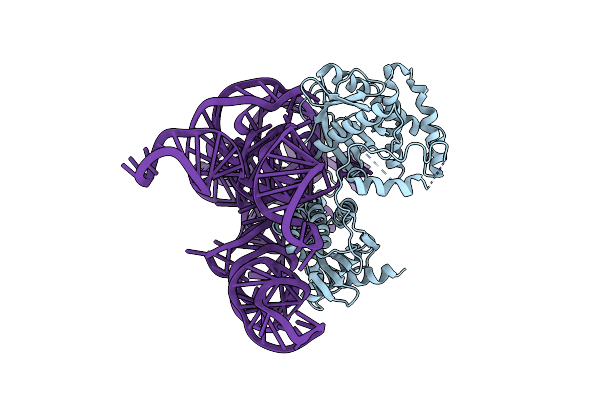
Cryo-Em Structure Of Shcas12K-Sgrna-Dsdna Ternary Complex (Type V-K Crispr-Associated Transposon)
Organism: Scytonema hofmannii
Method: ELECTRON MICROSCOPY Release Date: 2024-04-10 Classification: DNA BINDING PROTEIN |
 |
Cryo-Em Structure Of Cas12K-Sgrna Binary Complex (Type V-K Crispr-Associated Transposon)
Organism: Scytonema hofmannii
Method: ELECTRON MICROSCOPY Release Date: 2024-04-10 Classification: DNA BINDING PROTEIN |
 |
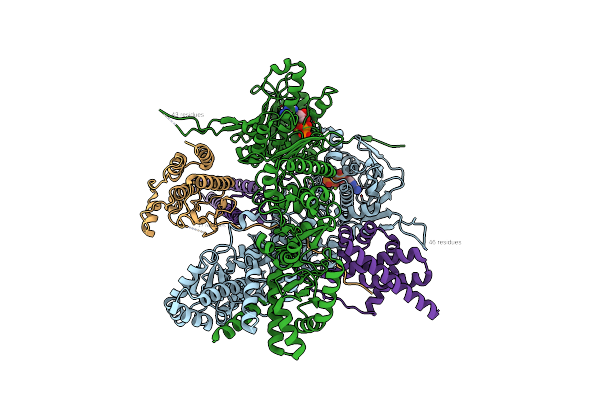
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: TRANSFERASE Ligands: ATP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: TRANSFERASE Ligands: ATP, MG |
 |
Organism: Prevotella buccae
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-02-20 Classification: HYDROLASE/RNA Ligands: CIT, CL, PG4, NA |
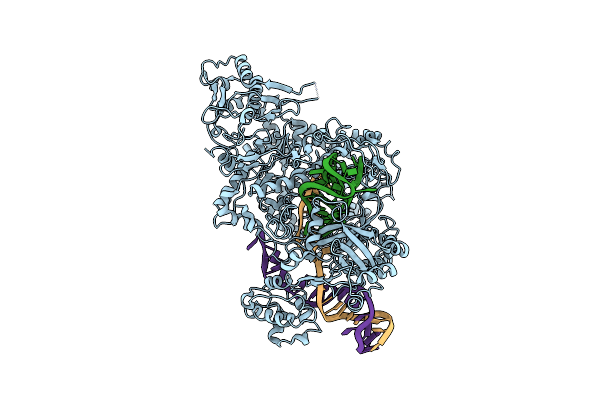 |
Organism: Francisella tularensis subsp. novicida u112
Method: ELECTRON MICROSCOPY Release Date: 2018-12-19 Classification: HYDROLASE |
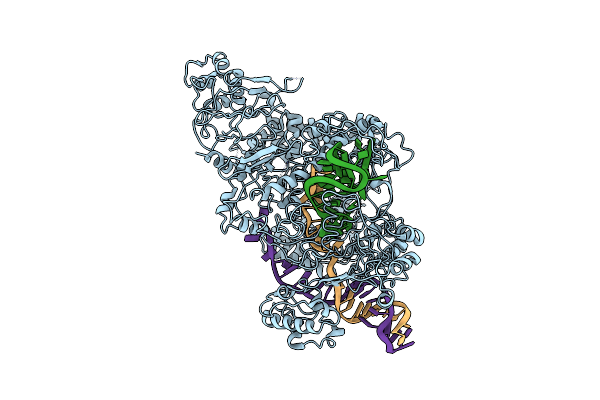 |
Organism: Francisella tularensis subsp. novicida (strain u112), Francisella tularensis subsp. novicida u112
Method: ELECTRON MICROSCOPY Release Date: 2018-12-19 Classification: HYDROLASE |
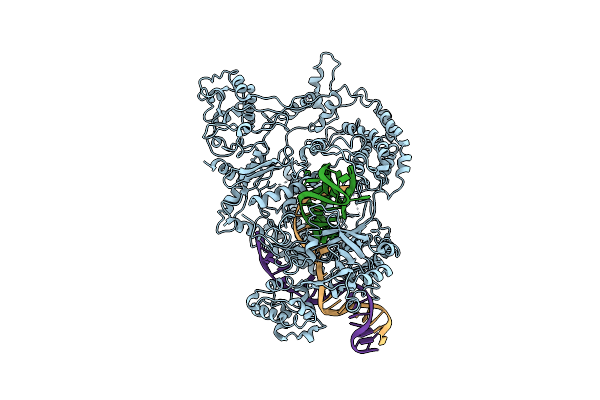 |
Organism: Francisella tularensis subsp. novicida (strain u112), Francisella tularensis subsp. novicida u112
Method: ELECTRON MICROSCOPY Release Date: 2018-12-19 Classification: HYDROLASE |
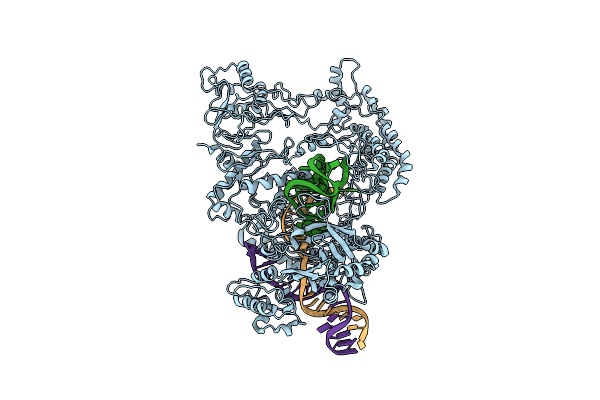 |
Organism: Francisella tularensis subsp. novicida u112
Method: ELECTRON MICROSCOPY Release Date: 2018-12-19 Classification: HYDROLASE |
 |
Organism: Francisella tularensis subsp. novicida u112
Method: ELECTRON MICROSCOPY Release Date: 2018-12-19 Classification: HYDROLASE Ligands: MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-09-13 Classification: CELL CYCLE Ligands: GOL |
 |
Crystal Structure Of The Mammalian Cytosolic Chaperonin Cct In Complex With Tubulin
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:5.50 Å Release Date: 2010-12-15 Classification: CHAPERONE |
 |
Structure Of Truncated Variant Of B.Subtilis Spp1 Phage G39P Helicase Loader/Inhibitor Protein
Organism: Bacillus phage spp1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2003-05-06 Classification: REPLICATION |

