Search Count: 25
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: CLR |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: A1HZ5, CLR |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: A1H0C, CLR |
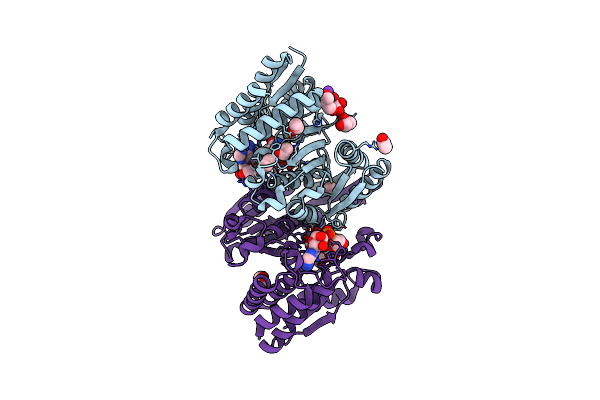 |
Structure Of The E270A Mutant Isopropylmalate Dehydrogenase From Thermus Thermophilus In Complex With Ipm, Mn And Nadh
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2014-11-12 Classification: OXIDOREDUCTASE Ligands: IPM, NAD, MN, K, EOH, GOL |
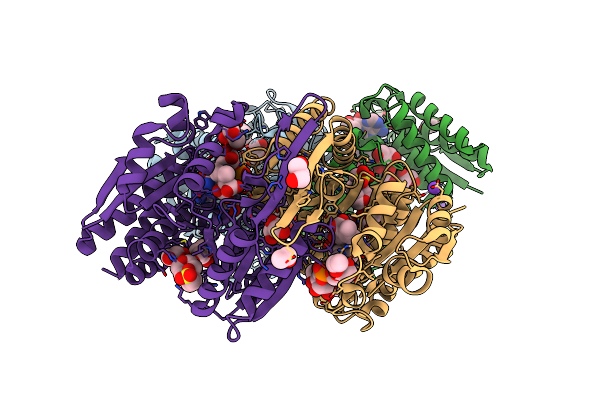 |
Structure Of Isopropylmalate Dehydrogenase From Thermus Thermophilus In Complex With Ipm, Mn And Nadh
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-06-13 Classification: OXIDOREDUCTASE Ligands: IPM, NAD, MPO, MN, K, GOL, EOH |
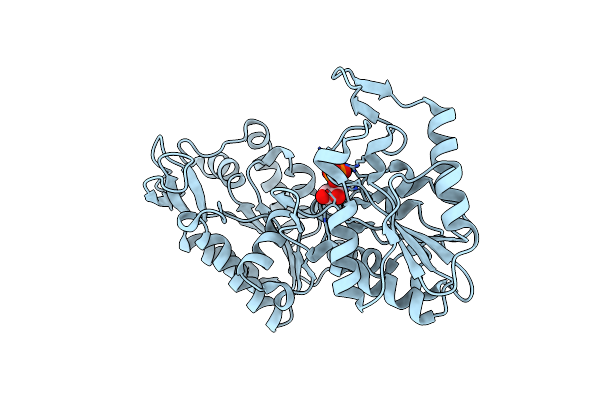 |
The Complete Reaction Cycle Of Human Phosphoglycerate Kinase: The Open Binary Complex With 3Pg
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2011-01-19 Classification: TRANSFERASE Ligands: 3PG |
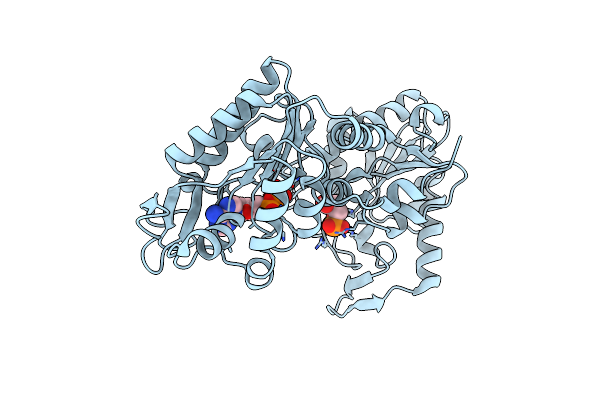 |
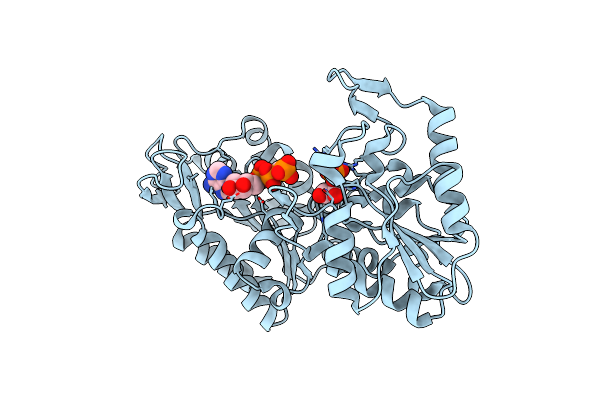
The Complete Reaction Cycle Of Human Phosphoglycerate Kinase: The Open Ternary Complex With 3Pg And Adp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2011-01-19 Classification: TRANSFERASE Ligands: 3PG, ADP |
 |
The Complete Reaction Cycle Of Human Phosphoglycerate Kinase: The Open Ternary Complex With 3Pg And Amp-Pnp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2011-01-19 Classification: TRANSFERASE Ligands: 3PG, ACP |
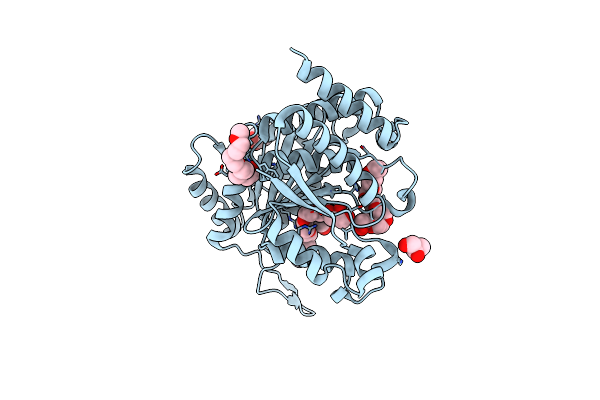 |
Structure Of Isopropylmalate Dehydrogenase From Thermus Thermophilus - Apo Enzyme
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2011-01-19 Classification: OXIDOREDUCTASE Ligands: TRS, 2PE, GOL |
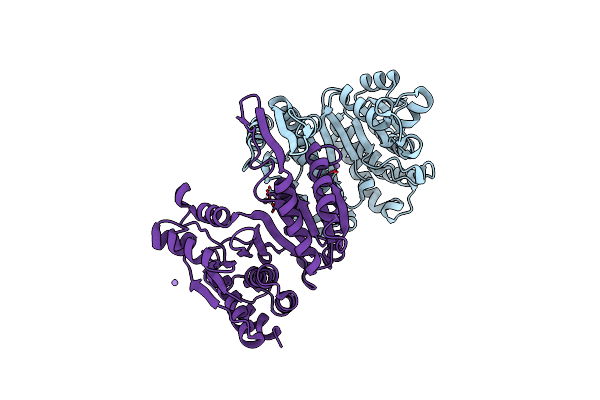 |
Structure Of Isopropylmalate Dehydrogenase From Thermus Thermophilus - Complex With Mn
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-01-19 Classification: OXIDOREDUCTASE Ligands: MN |
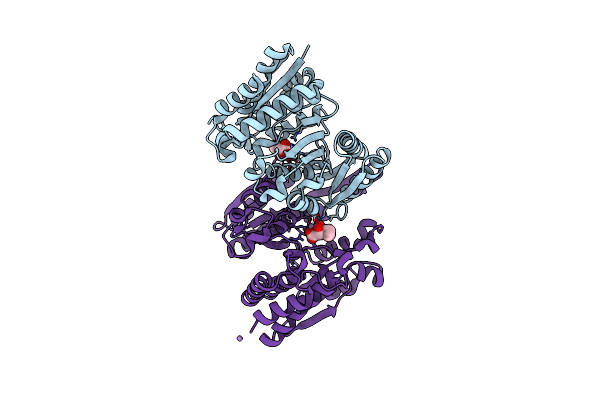 |
Structure Of Isopropylmalate Dehydrogenase From Thermus Thermophilus - Complex With Ipm And Mn
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2011-01-19 Classification: OXIDOREDUCTASE Ligands: IPM, MN |
 |
Structure Of Isopropylmalate Dehydrogenase From Thermus Thermophilus - Complex With Nadh And Mn
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-01-19 Classification: OXIDOREDUCTASE Ligands: BCN, NAD, MN |
 |
Crystal Structures Of The G81A Mutant Of The Active Chimera Of (S)-Mandelate Dehydrogenase And Its Complex With Two Of Its Substrates
Organism: Pseudomonas putida, Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2009-12-22 Classification: OXIDOREDUCTASE Ligands: FMN, MES |
 |
Organism: Paracoccus versutus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-12-30 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Crystal Structure Of An Electron Transfer Complex Between Aromatic Amine Dehydrogenase And Azurin From Alcaligenes Faecalis (Form 3)
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2006-11-21 Classification: OXIDOREDUCTASE/electron transport Ligands: CU |
 |
Crystal Structure Of An Electron Transfer Complex Between Aromatic Amine Dephydrogenase And Azurin From Alcaligenes Faecalis (Form 1)
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2006-11-21 Classification: OXIDOREDUCTASE/electron transport Ligands: CU |
 |
Crystal Structure Of An Electron Transfer Complex Between Aromatic Amine Dephydrogenase And Azurin From Alcaligenes Faecalis (Form 2)
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2006-11-21 Classification: OXIDOREDUCTASE/electron transport Ligands: CU |
 |
Crystal Structure Of The G81A Mutant Of The Active Chimera Of (S)-Mandelate Dehydrogenase
Organism: Pseudomonas putida, Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-07-11 Classification: OXIDOREDUCTASE Ligands: FMN, MES |
 |
Crystal Structure Of The G81A Mutant Of The Active Chimera Of (S)-Mandelate Dehydrogenase In Complex With Its Substrate 3-Indolelactate
Organism: Pseudomonas putida, Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-07-11 Classification: OXIDOREDUCTASE Ligands: FMN, MES, 3IL |
 |
Crystal Structure Of The G81A Mutant Of The Active Chimera Of (S)-Mandelate Dehydrogenase In Complex With Its Substrate 2-Hydroxyoctanoate
Organism: Pseudomonas putida, Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2006-07-11 Classification: OXIDOREDUCTASE Ligands: FMN, MES, HOC |

