Search Count: 959
 |
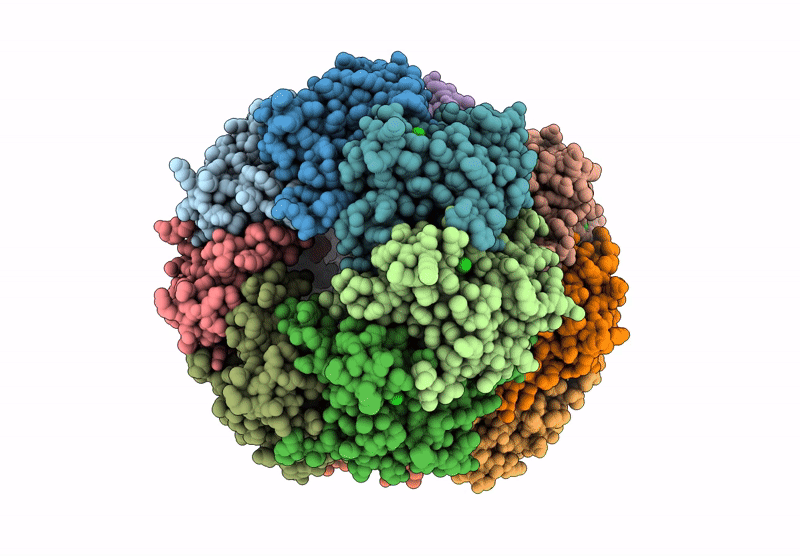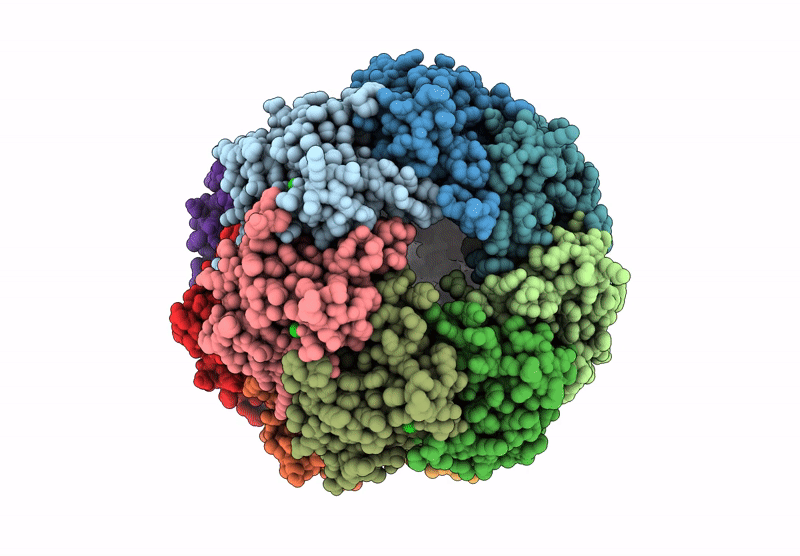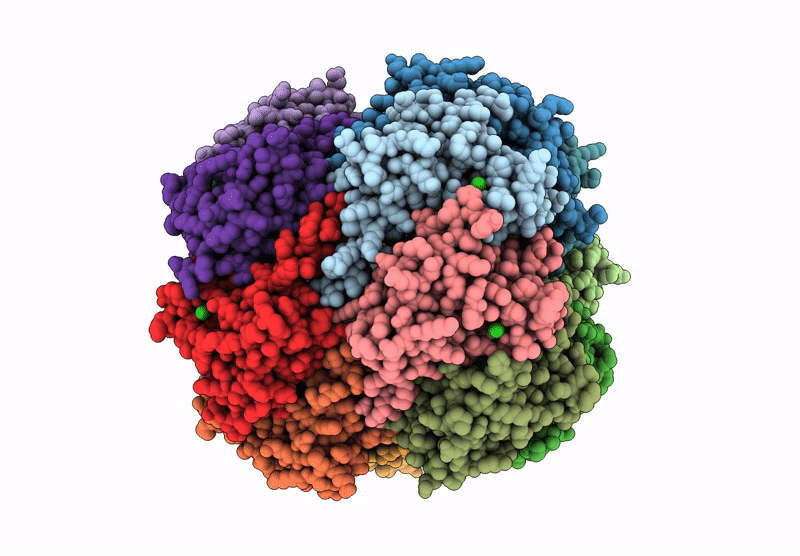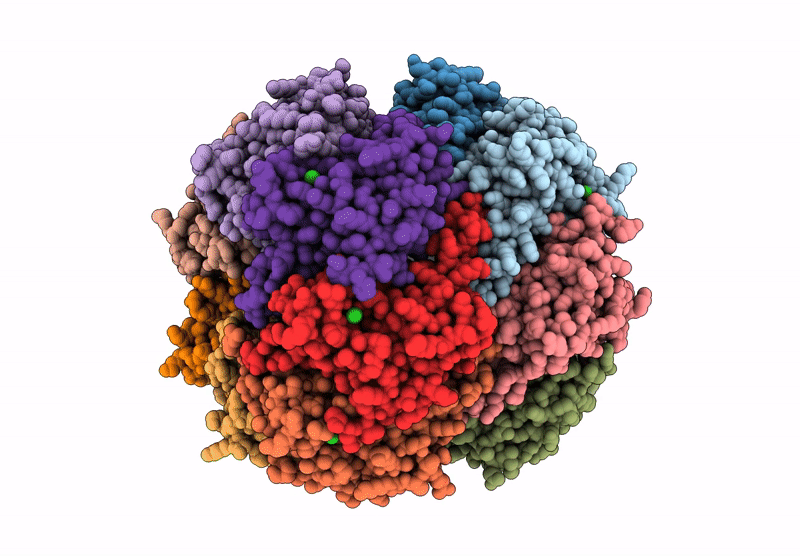
Structure Of The C-Terminal Half Of Lrrk2 Bound To Rn277 (Type-Ii Inhibitor)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: PROTEIN BINDING Ligands: A1A7Q |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2025-04-30 Classification: HYDROLASE Ligands: CL |
 |
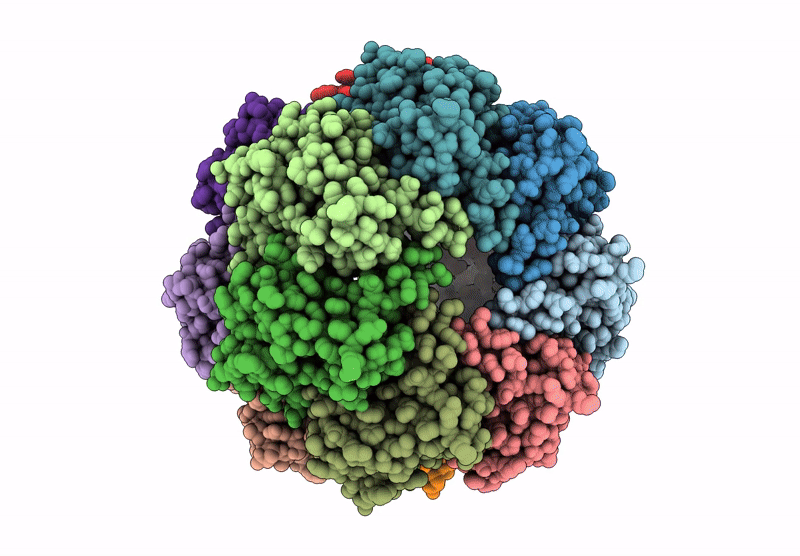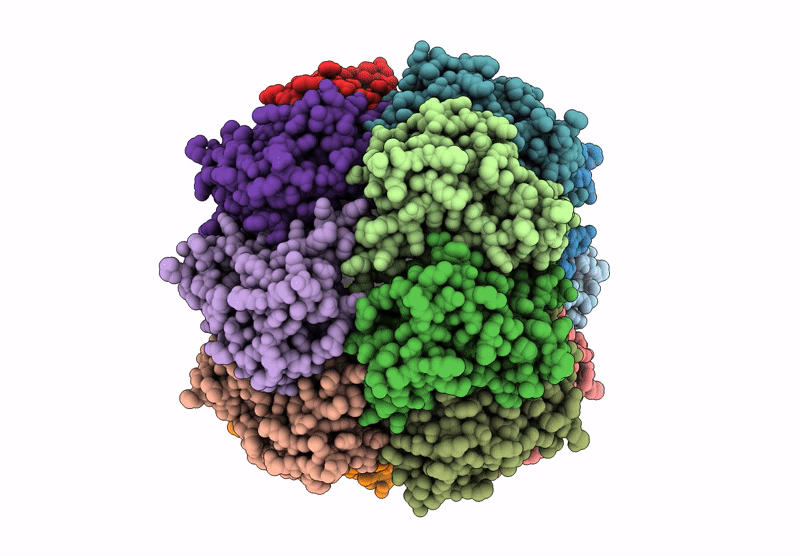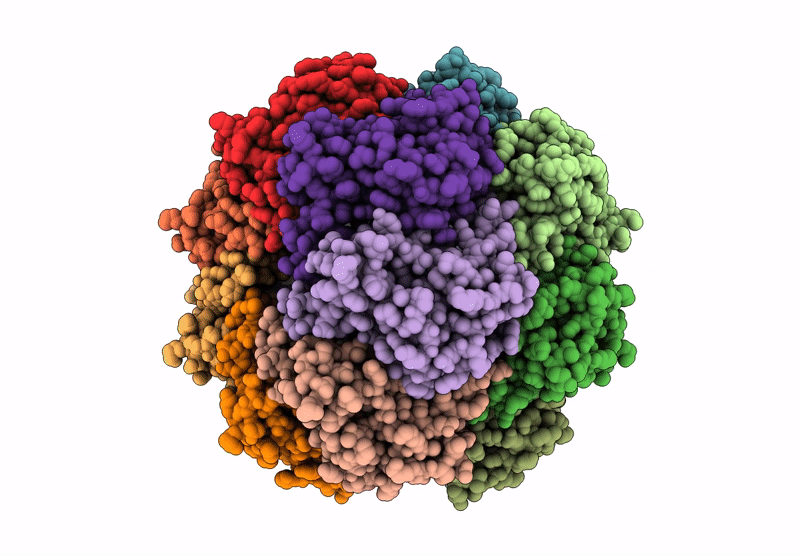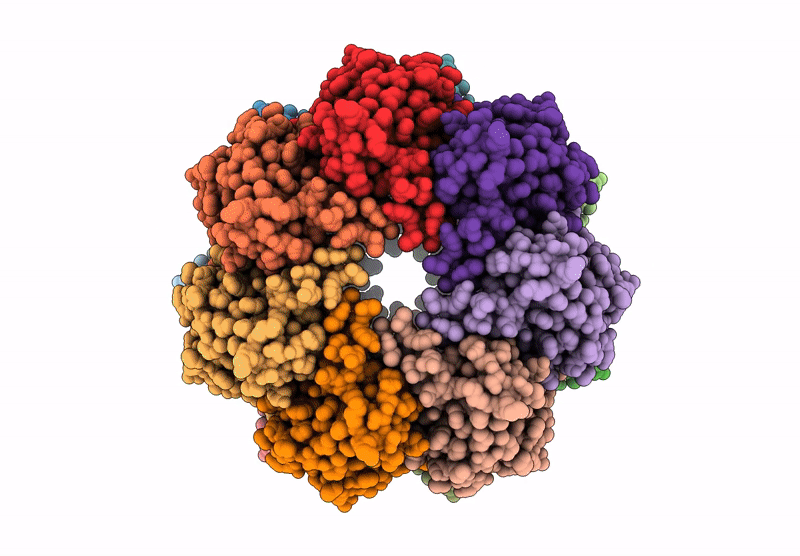
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2025-04-30 Classification: HYDROLASE Ligands: BO2, CL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: HYDROLASE |
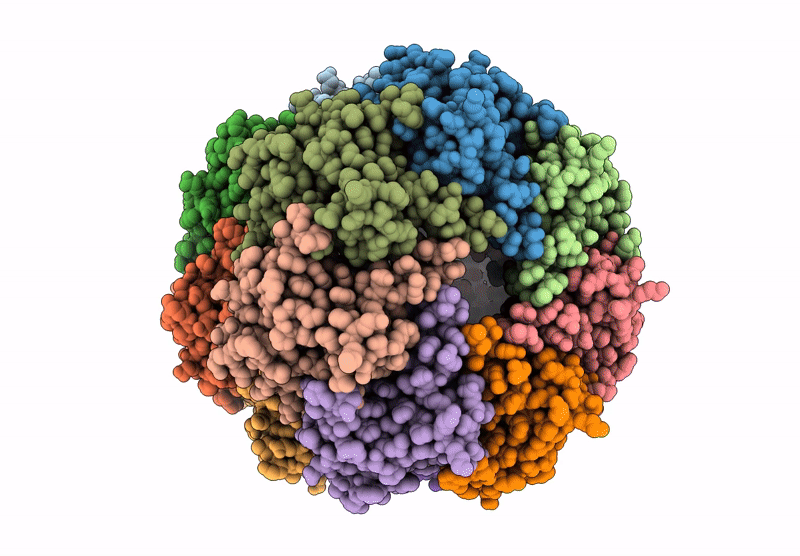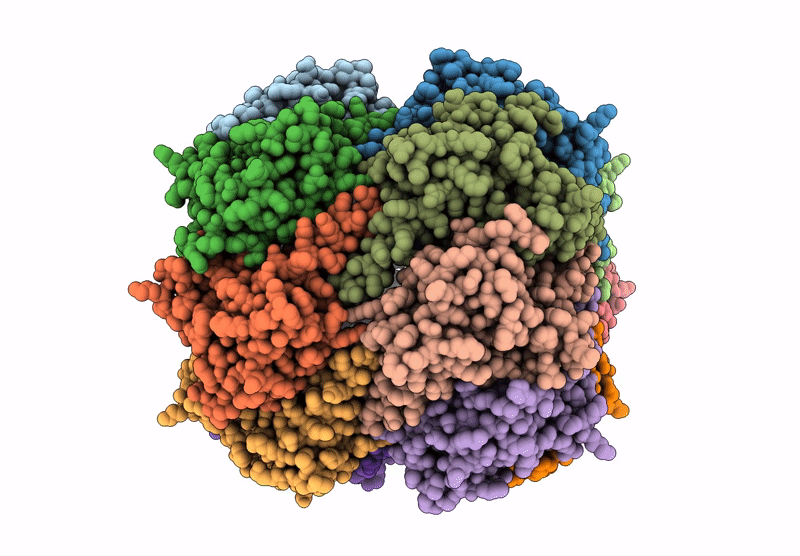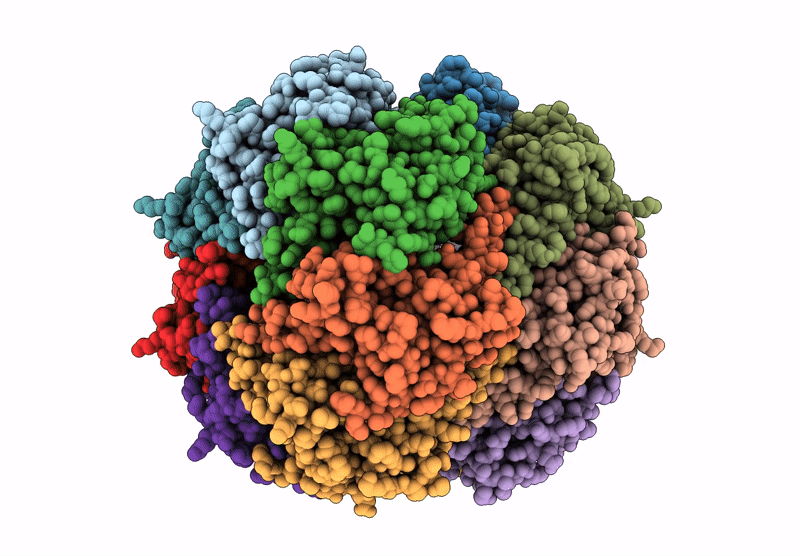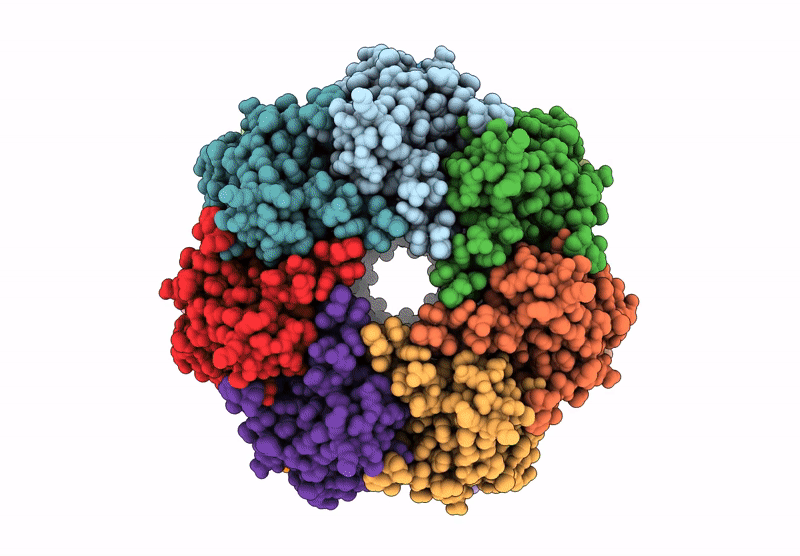 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: HYDROLASE Ligands: BO2 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-03-12 Classification: LIGASE Ligands: A1I41, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-03-05 Classification: OXIDOREDUCTASE Ligands: SO4, 9DJ, ACT, ORO, FMN |
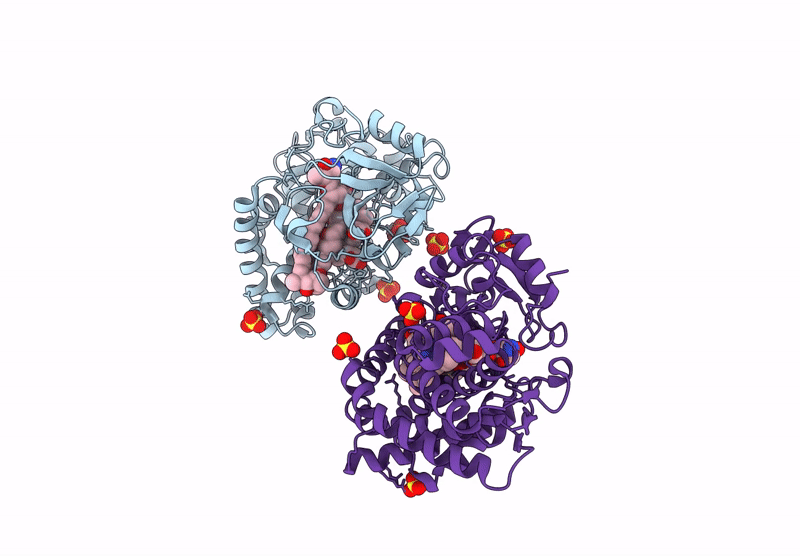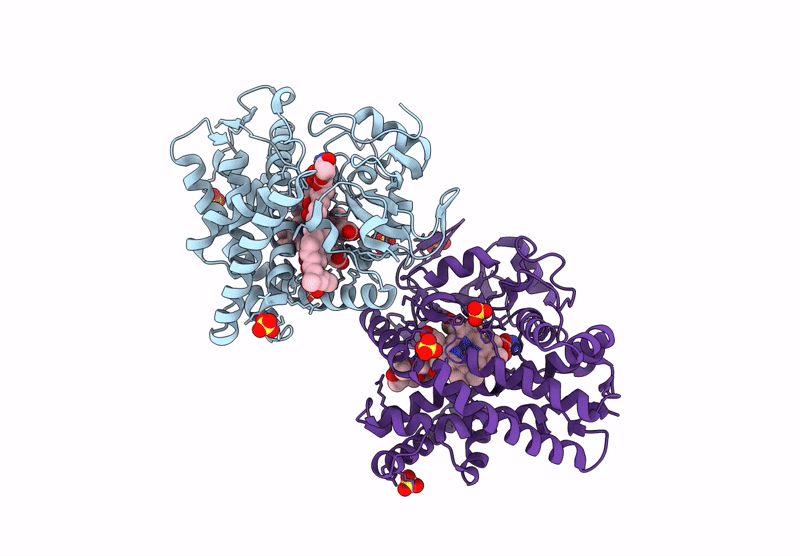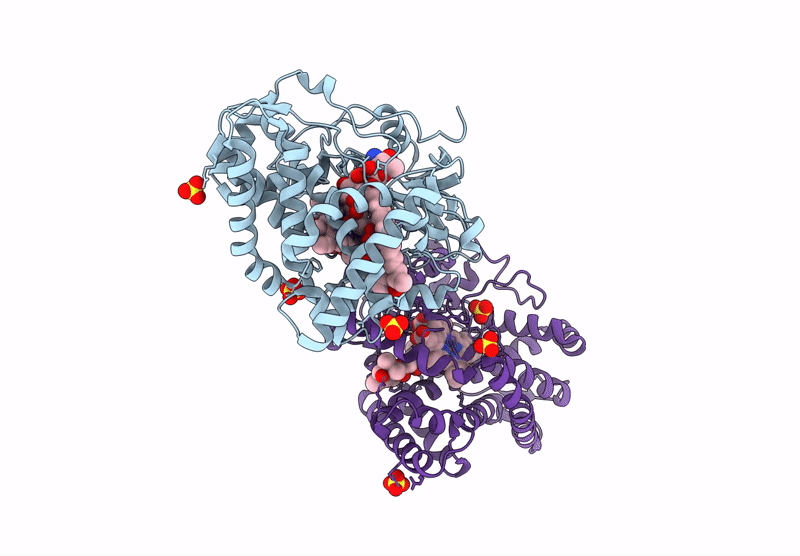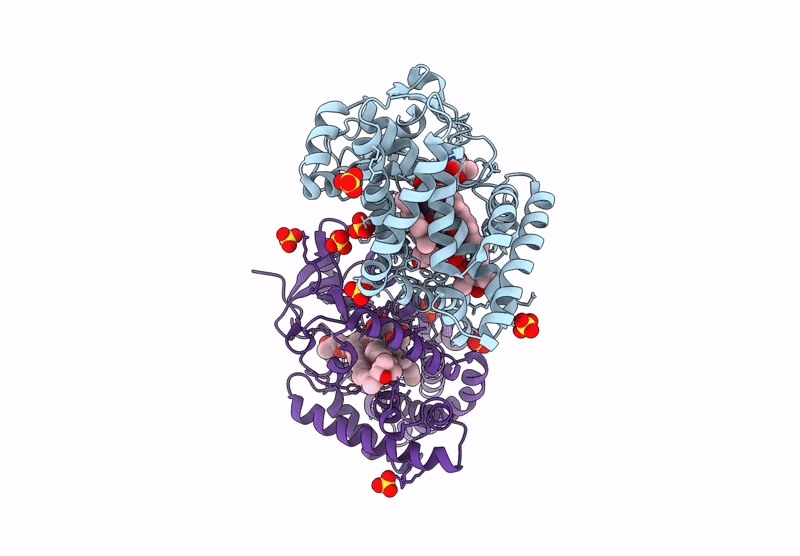 |
Organism: Streptomyces noursei
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: HEM, A1AZ7, SO4 |
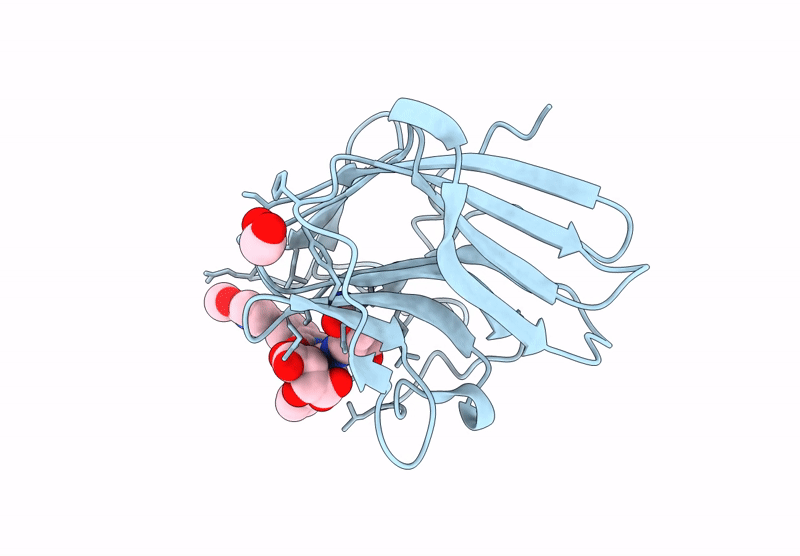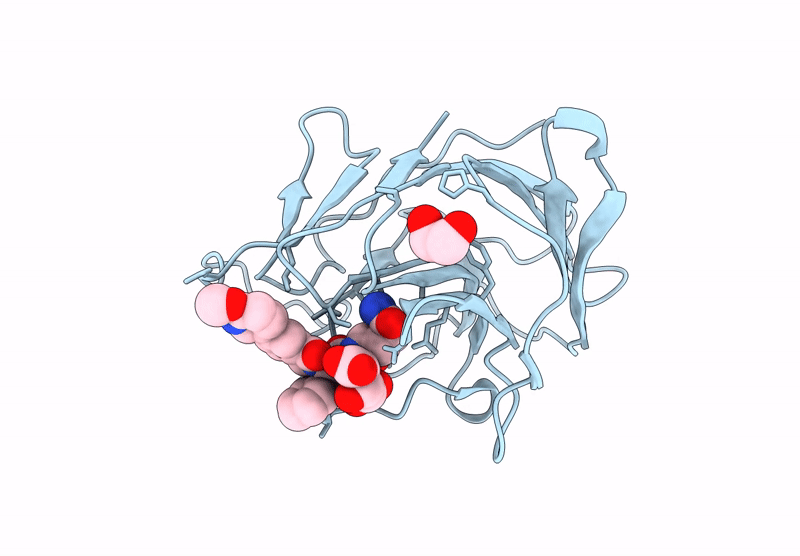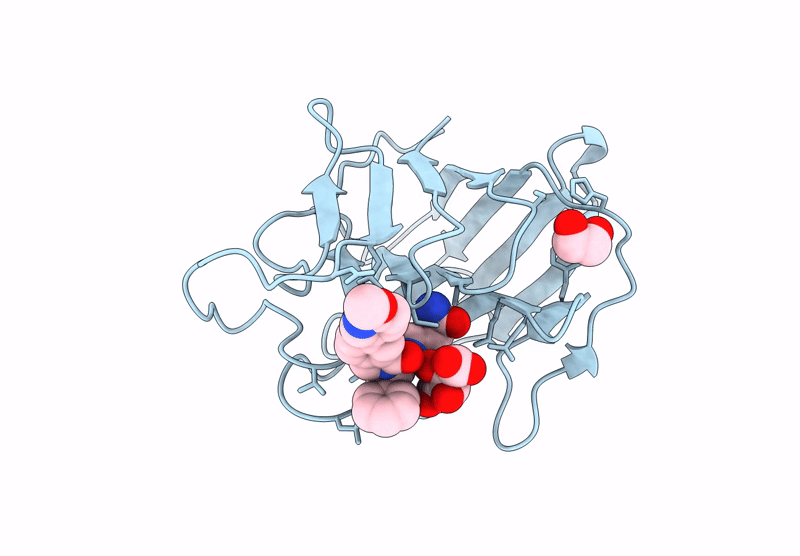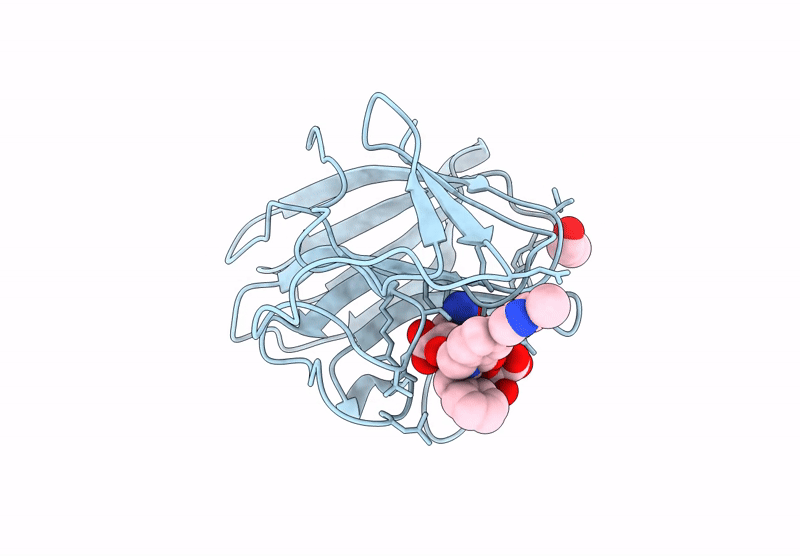 |
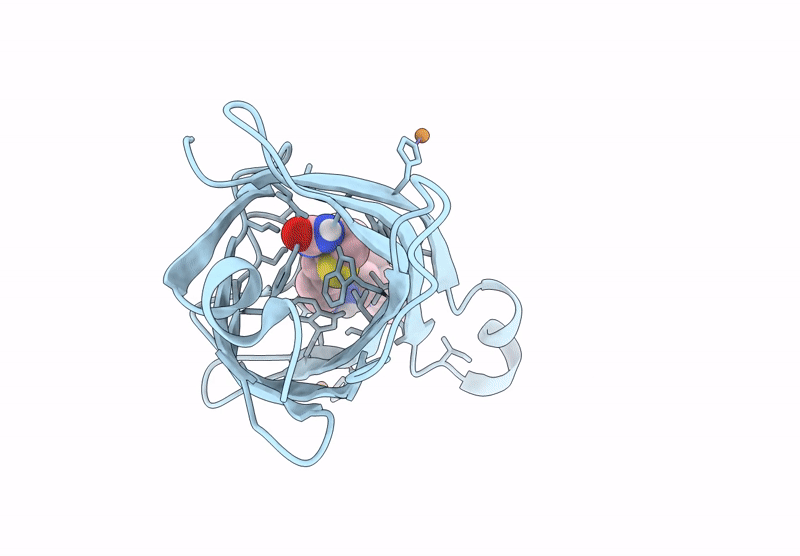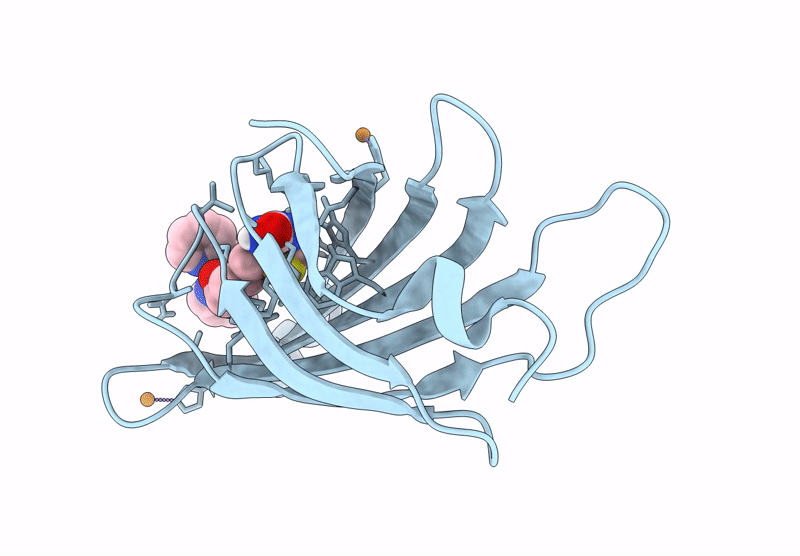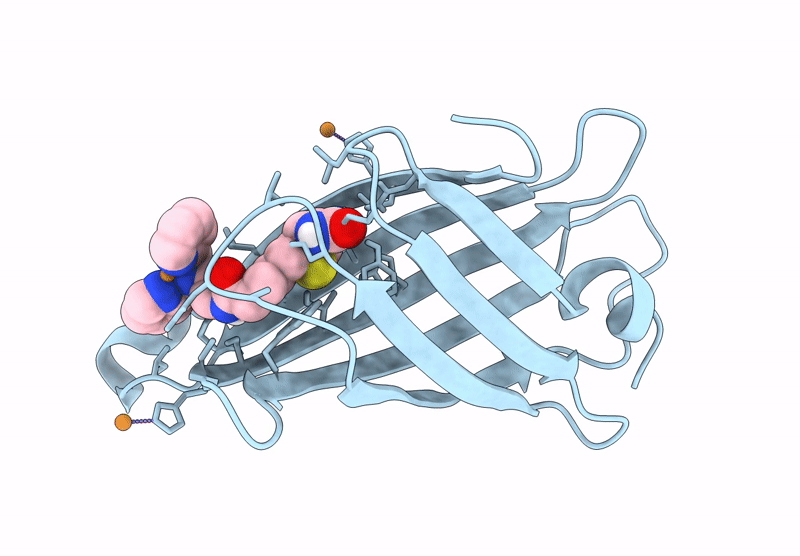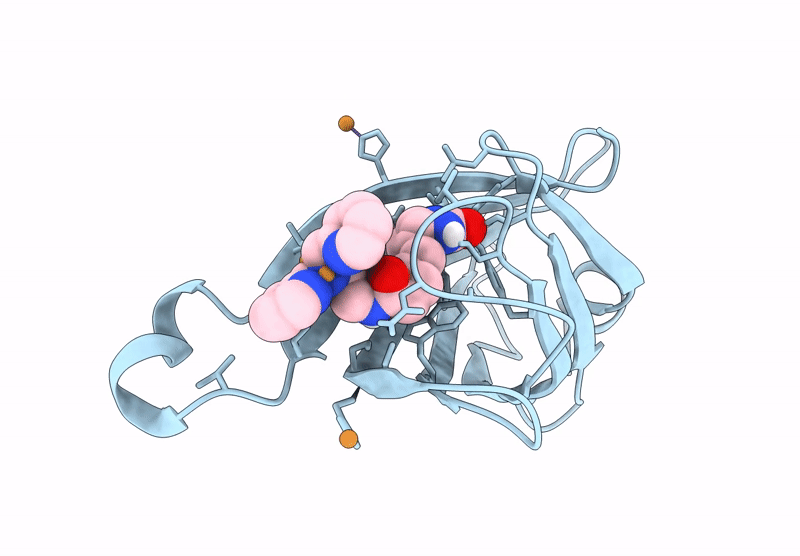
Crystal Structure Of Human Trim7 Pryspry Domain Bound To ((S)-2-(6-(6-Methoxypyridin-3-Yl)-1-Oxoisoindolin-2-Yl)-3-Phenylpropanoyl)-L-Glutamine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-01-15 Classification: LIGASE Ligands: TLA, A1IU9, EDO |
 |
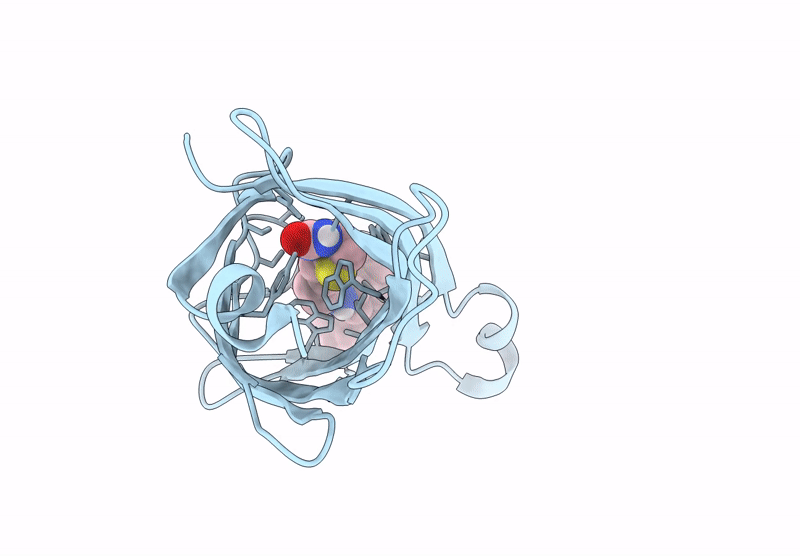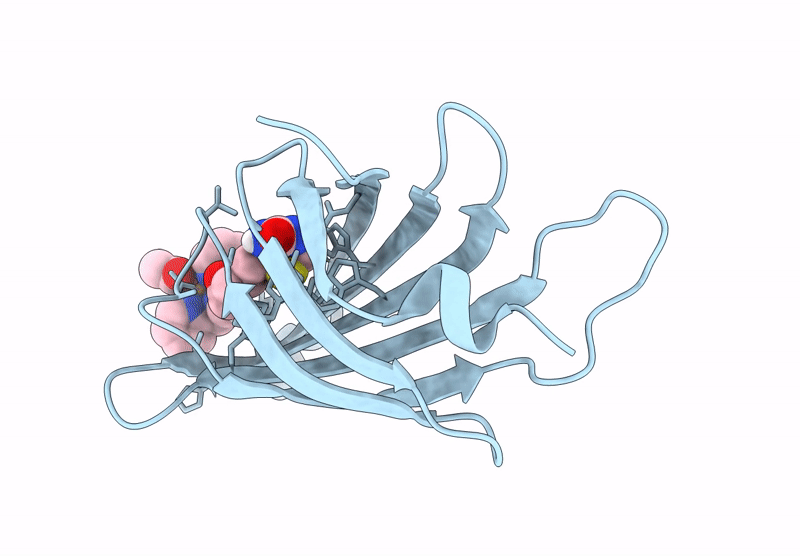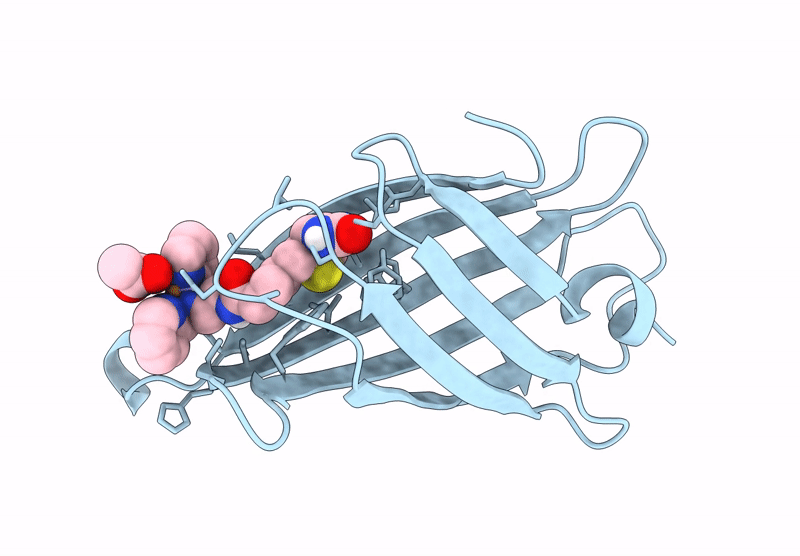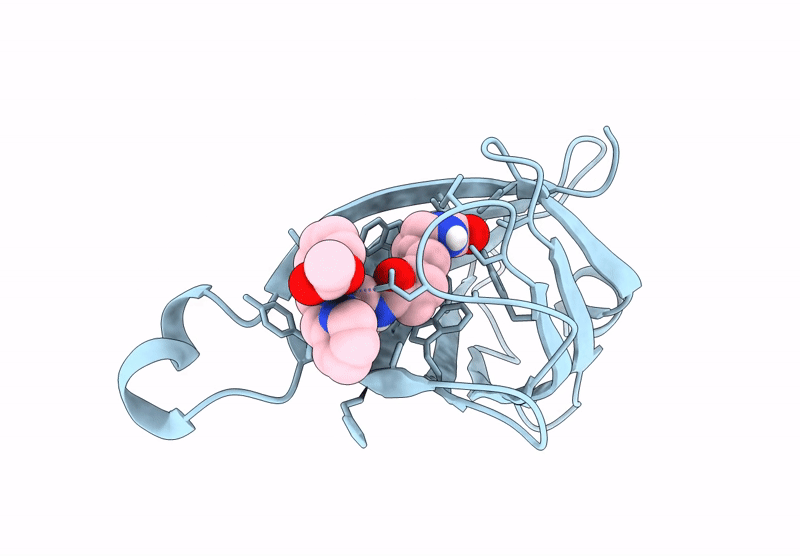
Streptavidin-E101Q-K121A Bound To Cu(Ii)-Biotin-Ethyl-Dipicolylamine Cofactor
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2024-12-11 Classification: METAL BINDING PROTEIN Ligands: QG7, CU |
 |
Streptavidin-E101Q-S112Y-K121A Bound To Cu(Ii)-Biotin-Ethyl-Dipicolylamine Cofactor
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-12-11 Classification: METAL BINDING PROTEIN Ligands: ACY, QG7, CU |
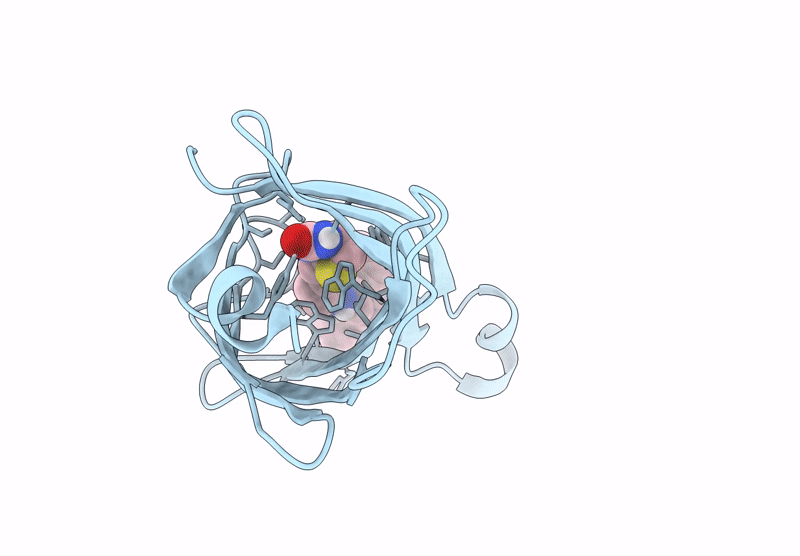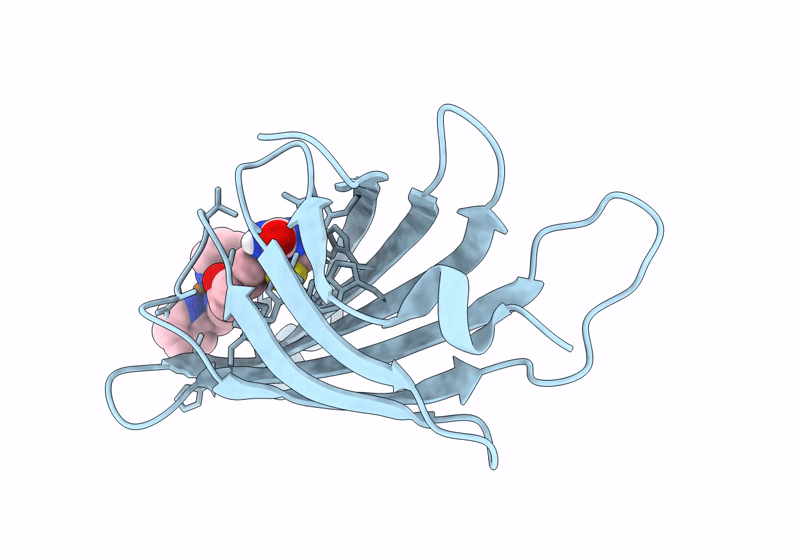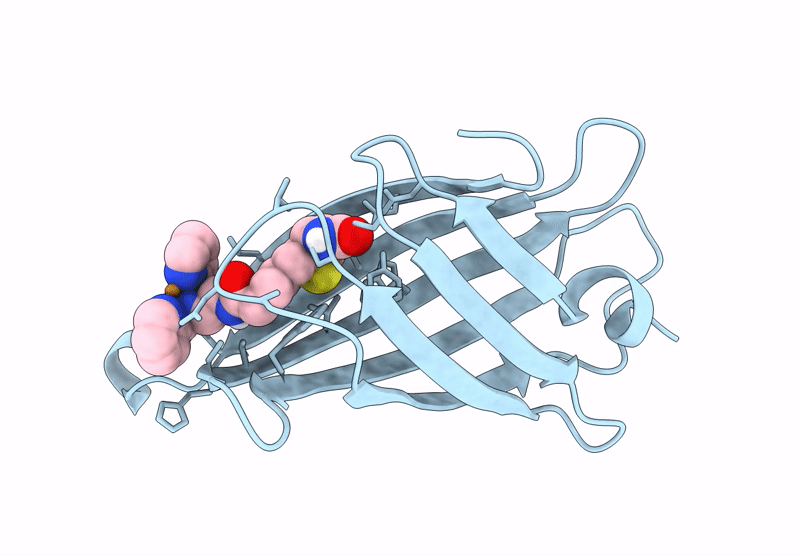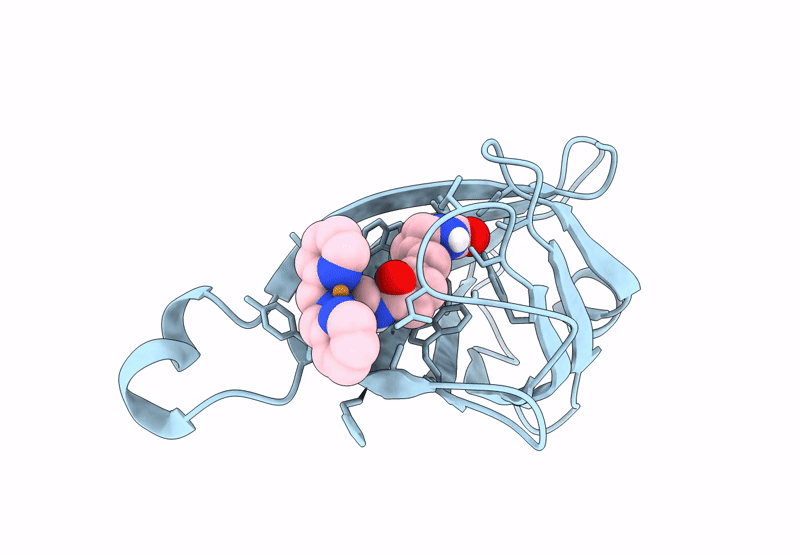 |
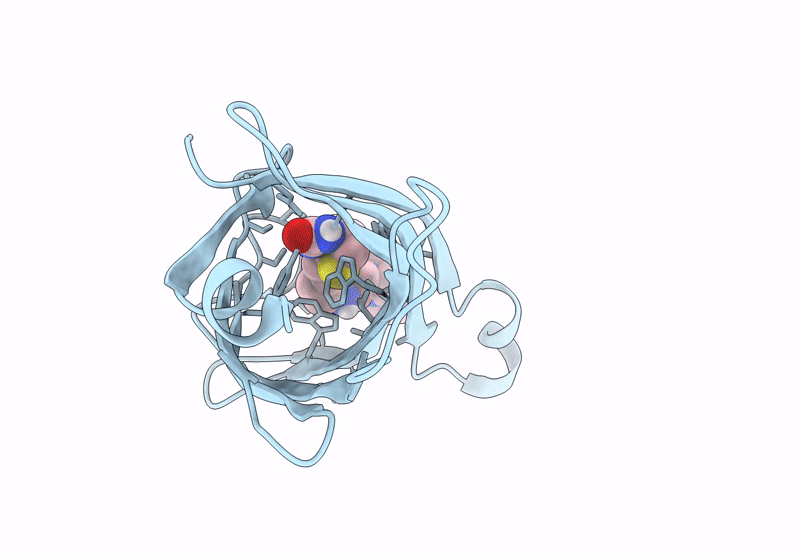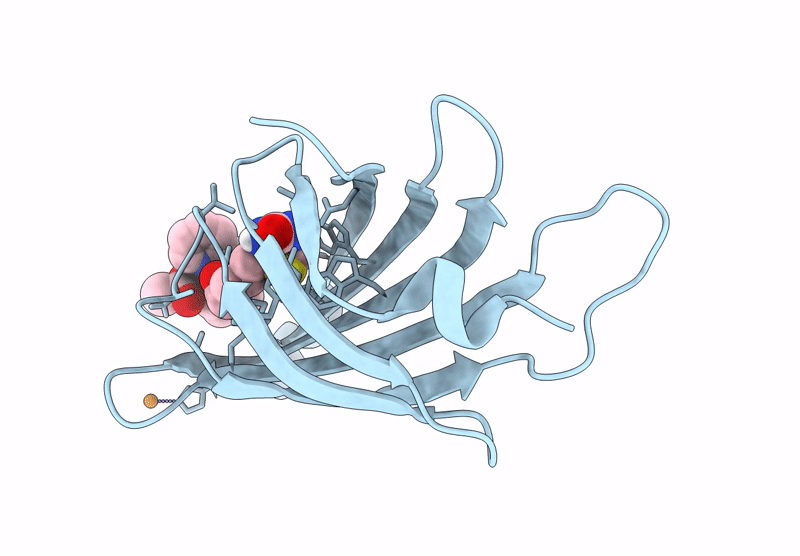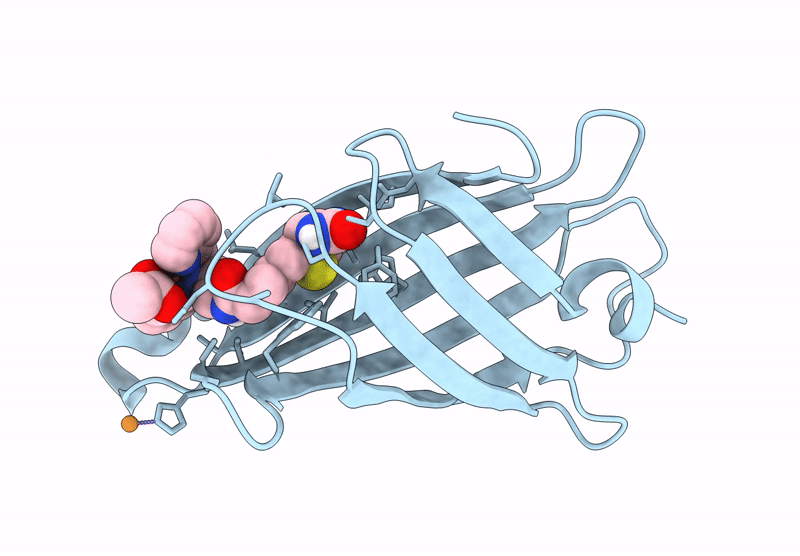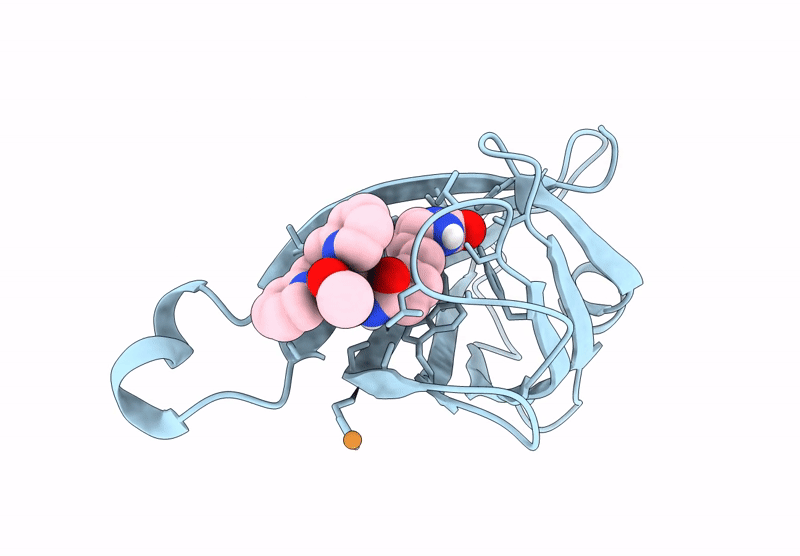
Streptavidin-E101Q-S112Y-K121A Bound To Cu(Ii)-Biotin-Ethyl-Dipicolylamine Cofactor, Oxidized By Hydrogen Peroxide
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-12-11 Classification: METAL BINDING PROTEIN Ligands: QG7, CU |
 |
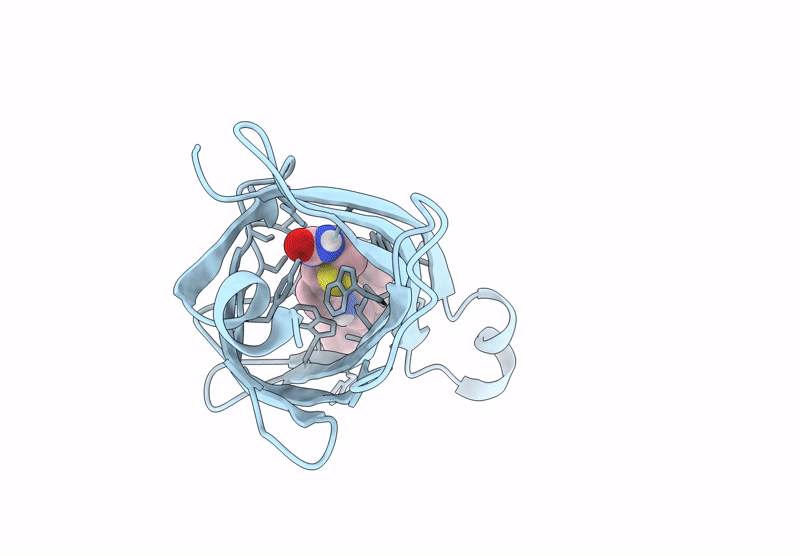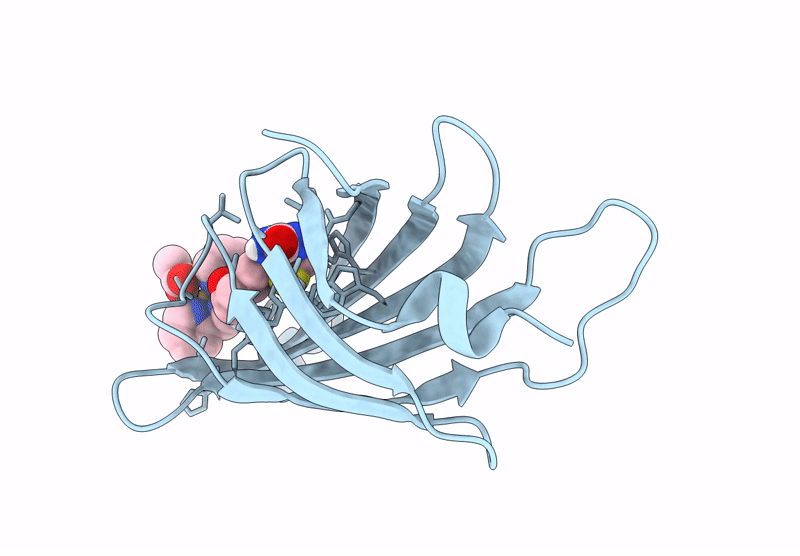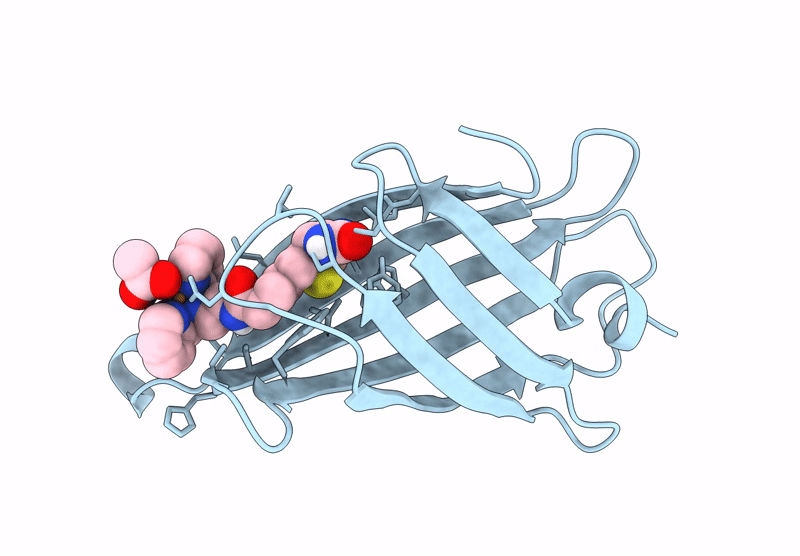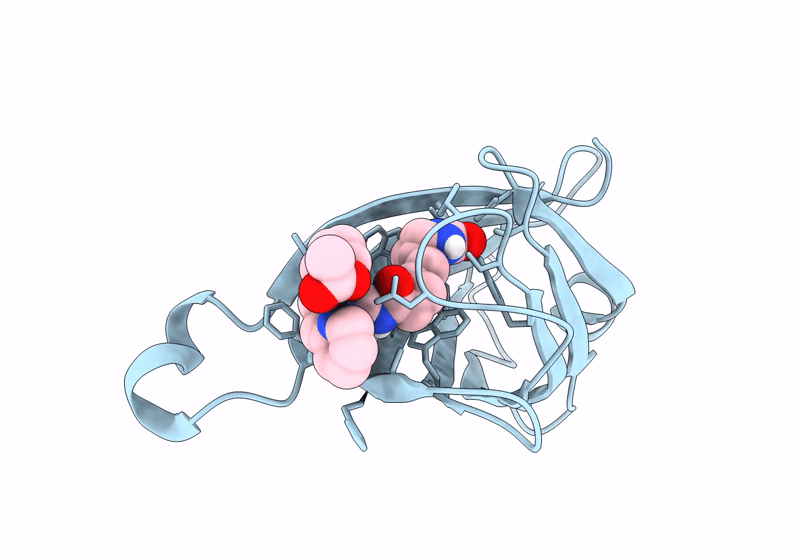
Streptavidin-E101Q-S112A-K121Y Bound To Cu(Ii)-Biotin-Ethyl-Dipicolylamine Cofactor
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2024-12-11 Classification: METAL BINDING PROTEIN Ligands: QG7, CU, ACY |
 |
Streptavidin-E101Q-S112F-K121A Bound To Cu(Ii)-Biotin-Ethyl-Dipicolylamine Cofactor
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-12-11 Classification: METAL BINDING PROTEIN Ligands: QG7, ACY, CU |
 |
Pandda Analysis Group Deposition -- Crystal Structure Of Trim21 In Complex With Z111634612
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2024-11-27 Classification: LIGASE Ligands: EDO, A1BE6, EPE, SO4 |
 |
Pandda Analysis Group Deposition -- Crystal Structure Of Trim21 In Complex With Z45705015
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2024-11-27 Classification: LIGASE Ligands: S9S, EPE, EDO, SO4 |
 |
Pandda Analysis Group Deposition -- Crystal Structure Of Trim21 In Complex With Z1079512010
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2024-11-27 Classification: LIGASE Ligands: EDO, UWS, SO4 |
 |
Pandda Analysis Group Deposition -- Crystal Structure Of Trim21 In Complex With Z1401276297
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2024-11-27 Classification: LIGASE Ligands: EDO, U0S, SO4 |
 |
Pandda Analysis Group Deposition -- Crystal Structure Of Trim21 In Complex With Z1696844792
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-11-27 Classification: LIGASE Ligands: UXG, EDO, SO4 |
 |
Pandda Analysis Group Deposition -- Crystal Structure Of Trim21 In Complex With Z2092555279
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2024-11-27 Classification: LIGASE Ligands: EDO, SO4, A1BE7 |

