Search Count: 55
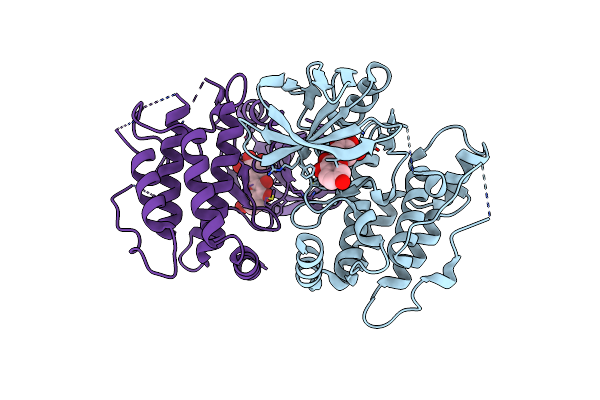 |
Crystal Structure Of The Mouse Rip3 Kinase Domain In Complexed With Robinetin
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2025-05-07 Classification: TRANSFERASE Ligands: LKR |
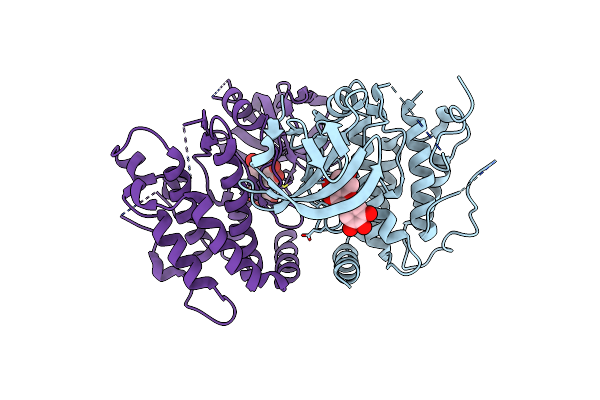 |
Crystal Structure Of The Mouse Rip3 Kinase Domain In Complexed With Tricetin
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2025-05-07 Classification: TRANSFERASE Ligands: MYF |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: SPLICING/RNA Ligands: M7M, MG, GTP, IHP, ZN |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: SPLICING/RNA Ligands: M7M, MG, GTP, ZN |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: SPLICING/RNA Ligands: M7M, GTP, ZN |
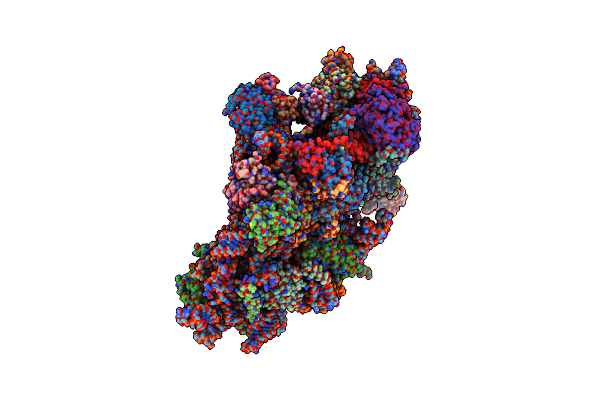 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: RIBOSOME Ligands: MG, ZN |
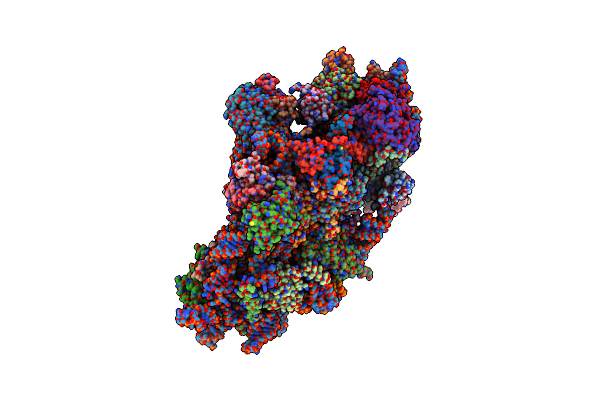 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:3.22 Å Release Date: 2024-03-20 Classification: OXIDOREDUCTASE Ligands: SO4, GOL |
 |
Leucine Dehydrogenase Structure In Ternary Complex With Nad+ From Bacillus Thuringiensis
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:3.52 Å Release Date: 2024-03-20 Classification: OXIDOREDUCTASE Ligands: NAD, SO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-02-28 Classification: CELL CYCLE Ligands: NAG, MAN |
 |
Crystal Structure Of Glucokinase (Hexokinase 4) Complexed With Ligand Diethyl ({2-[3-(4-Methanesulfonylpheno Xy)-5-{[(2S)-1-Methoxypropan-2-Yl]Oxy}Benzamido]-1,3-Thiaz Ol-4-Yl}Methyl)Phosphonate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-03-02 Classification: TRANSFERASE Ligands: GLC, NA, G2T, EDO |
 |
Crystal Structure Of Glucokinase (Hexokinase 4) Complexed With Ligand Aka Diethyl {[3-(3-{[5-(Azetidine-1-Carbon Yl)Pyrazin-2-Yl]Oxy}-5-(Propan-2-Yloxy)Benzamido)-1H- Pyrazol-1-Yl]Methyl}Phosphonate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-03-02 Classification: TRANSFERASE Ligands: G1S, GLC |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-03-04 Classification: PROTEIN TRANSPORT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-10-16 Classification: PROTEIN TRANSPORT |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2019-03-13 Classification: DE NOVO PROTEIN |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-02-10 Classification: METAL TRANSPORT Ligands: AU |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-02-10 Classification: METAL TRANSPORT |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-02-10 Classification: METAL TRANSPORT Ligands: AU |
 |
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-07-23 Classification: OXIDOREDUCTASE |

