Search Count: 62
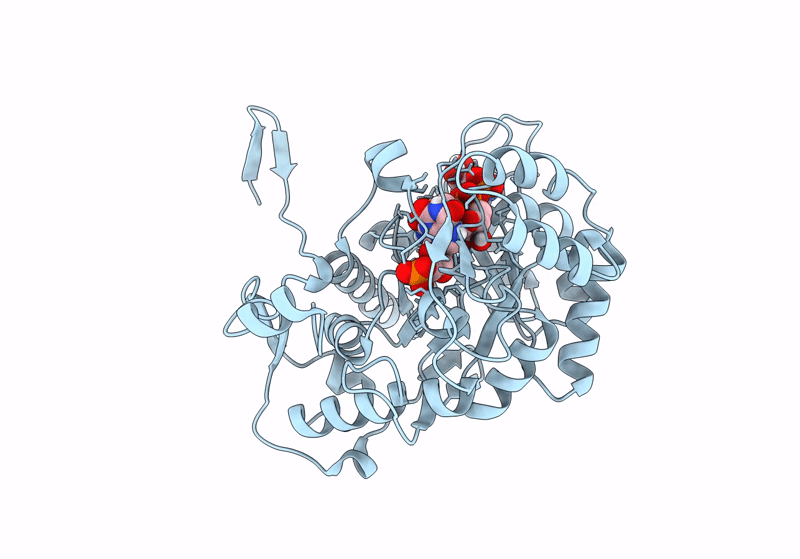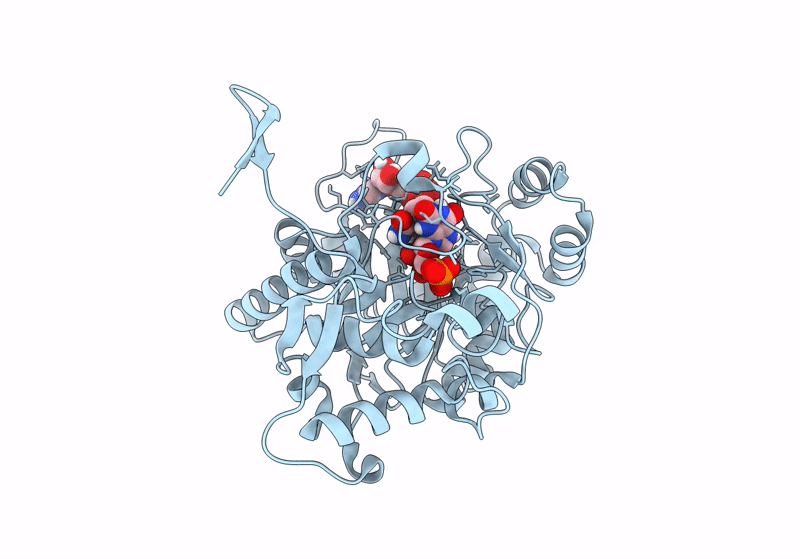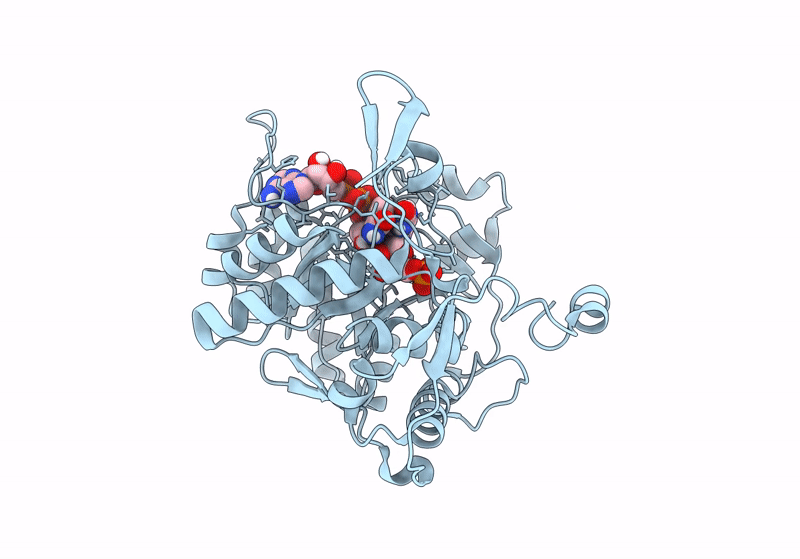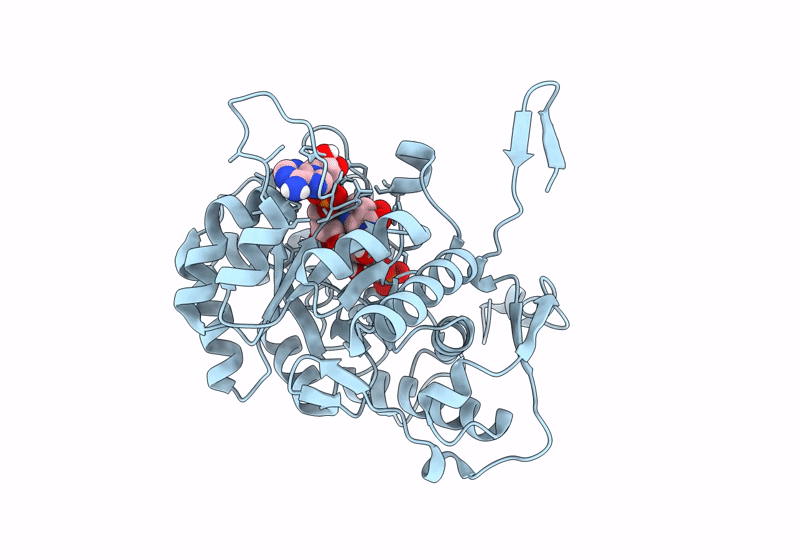 |
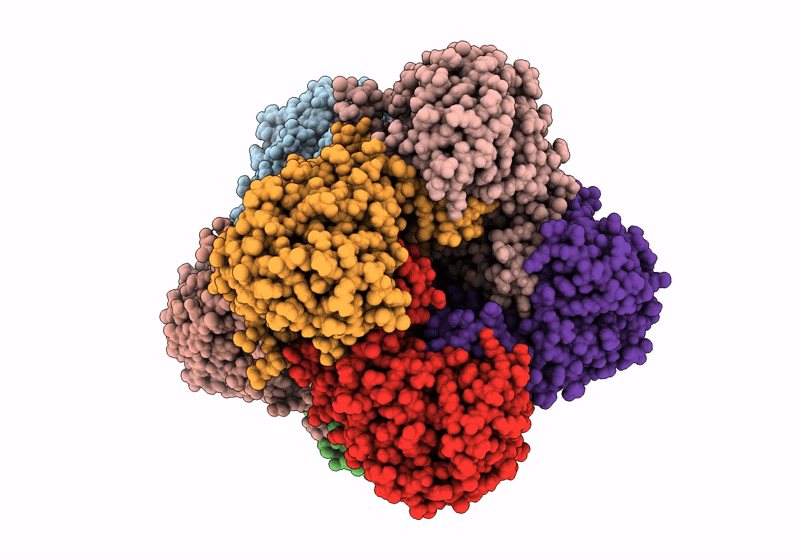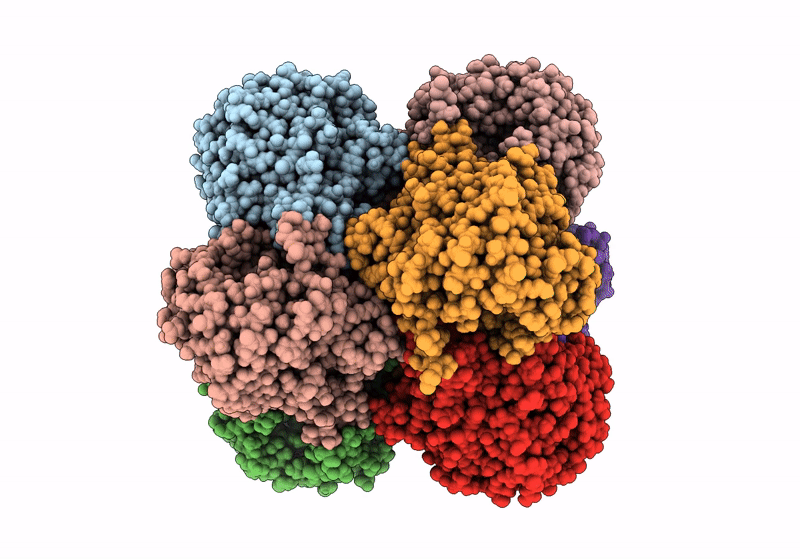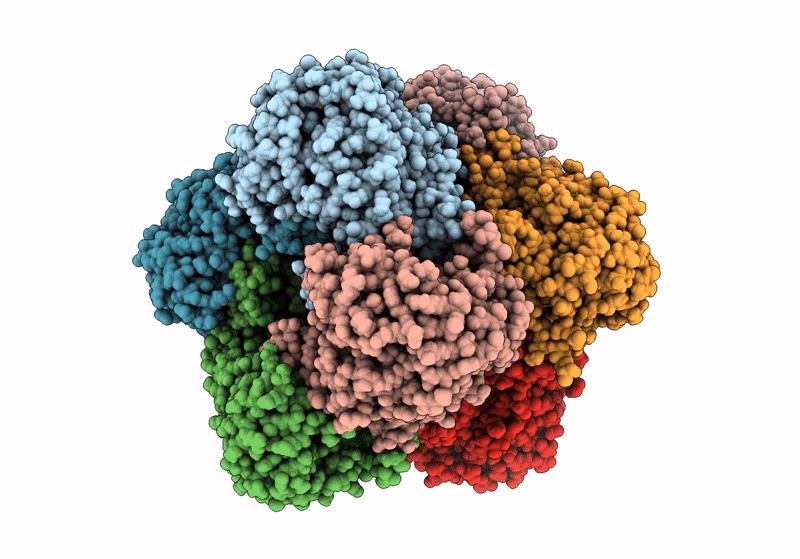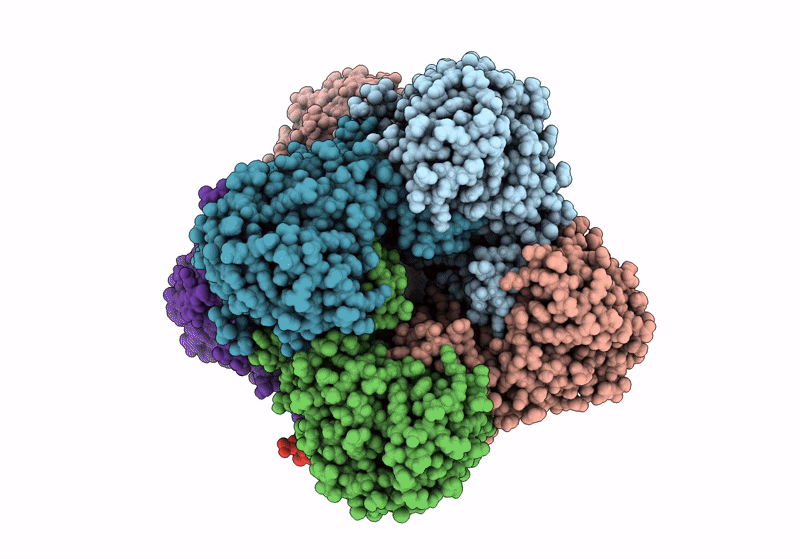
Structure A Catalytically Inactive Mutant Of The Imp Dehydrogenase Related Protein Guab3 From Synechocystis Pcc 6803
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2024-03-27 Classification: BIOSYNTHETIC PROTEIN Ligands: XMP, NAD |
 |
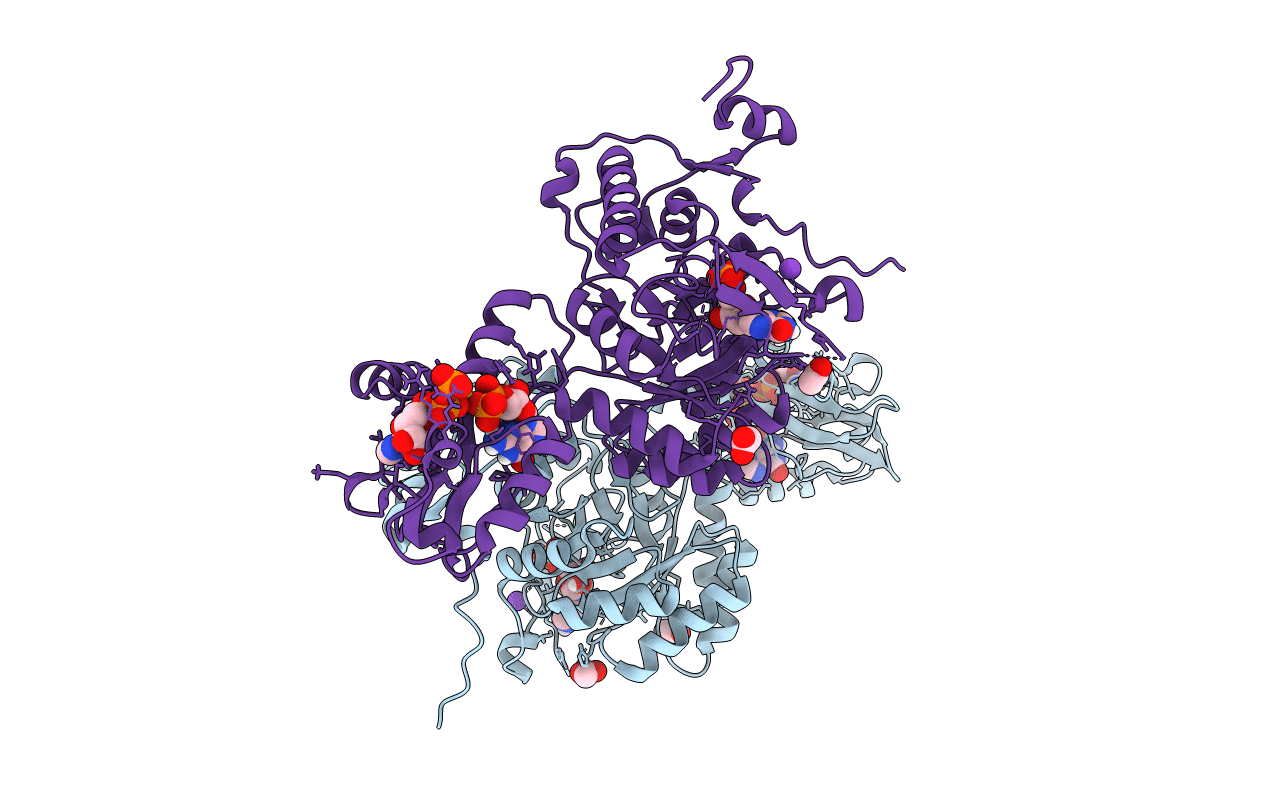
Structure Of The Imp Dehydrogenase Related Protein Guab3 From Synechocystis Pcc 6803
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2024-03-27 Classification: BIOSYNTHETIC PROTEIN Ligands: IMP, XMP |
 |
Crystal Structure Of Pseudomonas Aeruginosa Guab (Imp Dehydrogenase) Bound To Atp And Gdp At 1.65A Resolution
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-07-06 Classification: OXIDOREDUCTASE Ligands: ATP, GDP, IMP, MG, K, ACT |
 |
Crystal Structure Of Streptomyces Coelicolor Guab (Imp Dehydrogenase) Bound To Atp And Ppgpp At 2.0 A Resolution
Organism: Streptomyces coelicolor a3(2)
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2022-05-11 Classification: OXIDOREDUCTASE Ligands: ATP, G4P, MG |
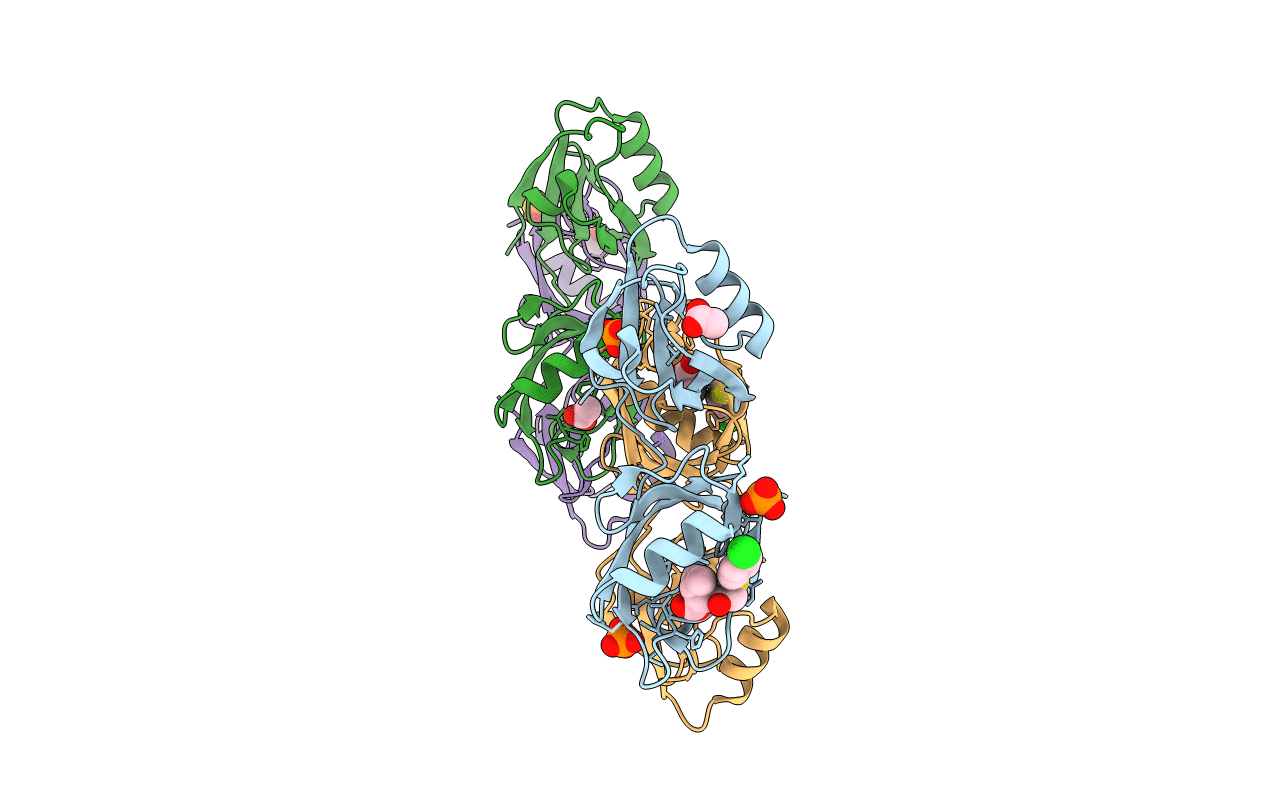 |
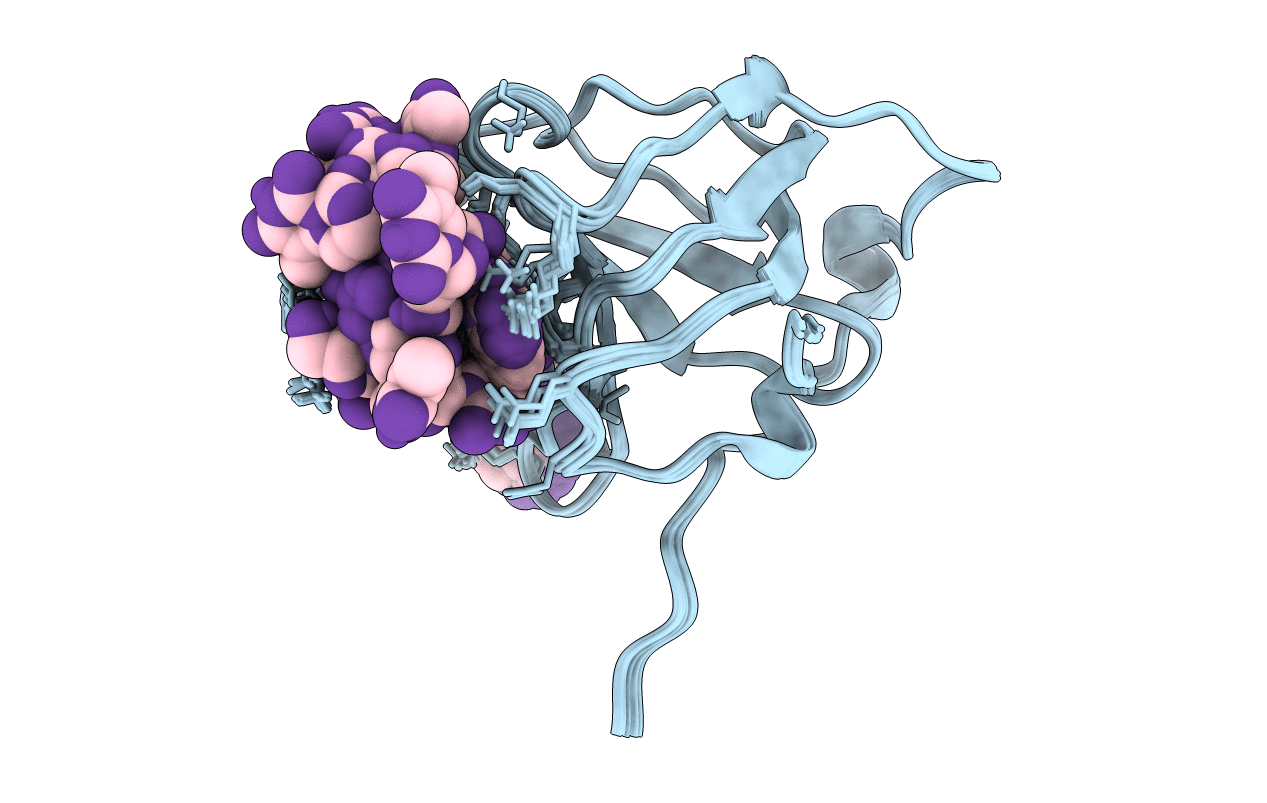
Crystal Structure Of The Pdz Tandem Of Syntenin In Complex With Fragment C58
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-02-03 Classification: SIGNALING PROTEIN Ligands: PO4, JVK, ACT |
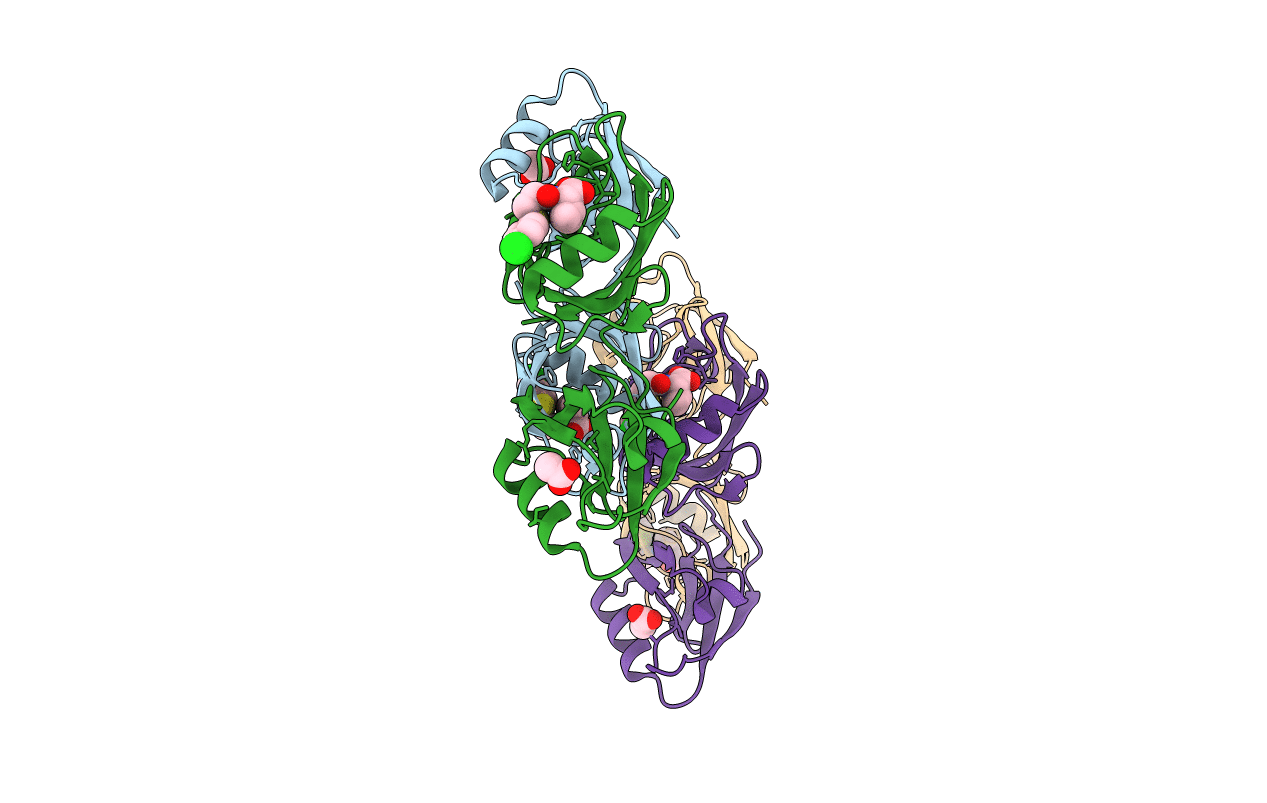 |
Crystal Structure Of The Pdz Tandem Of Syntenin In Complex With Fragment F13
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-02-03 Classification: SIGNALING PROTEIN Ligands: K7Z, ACT |
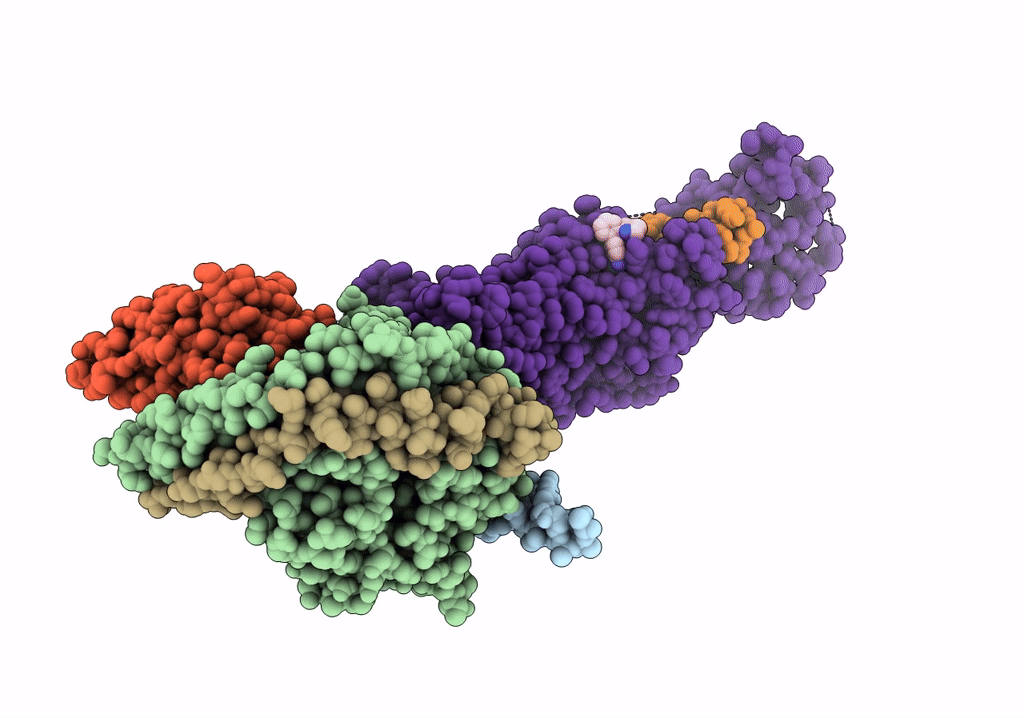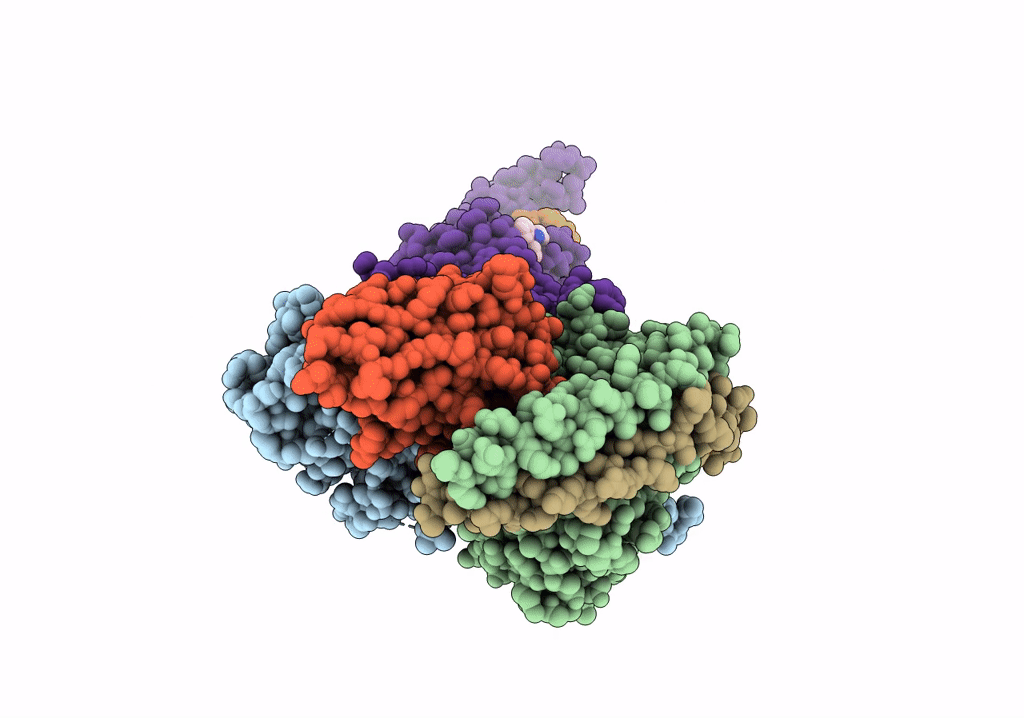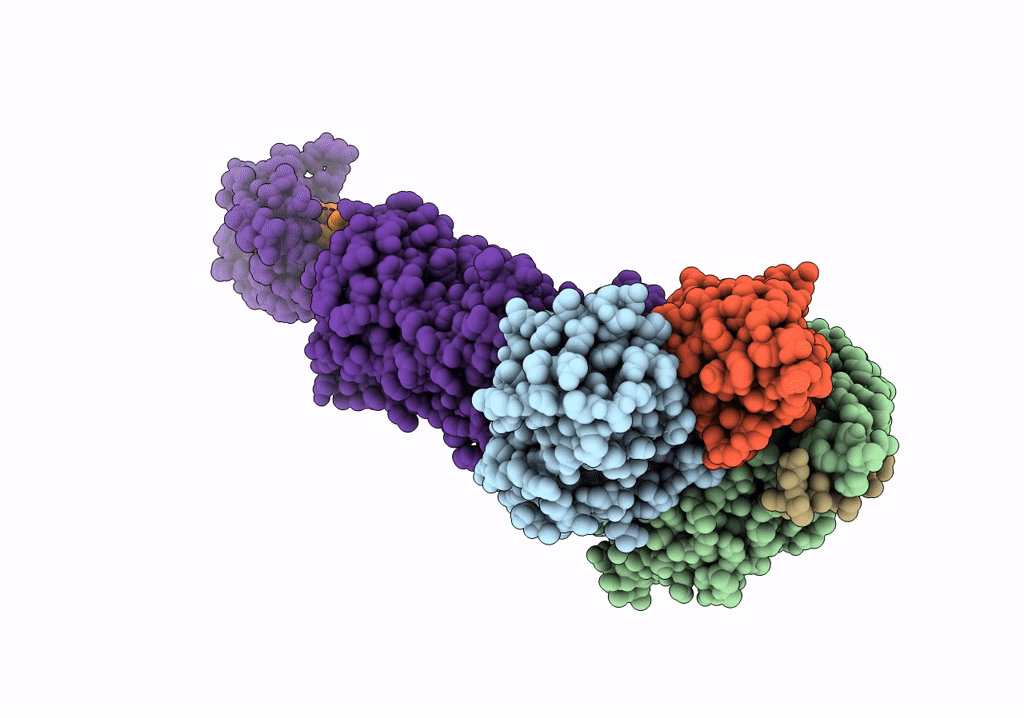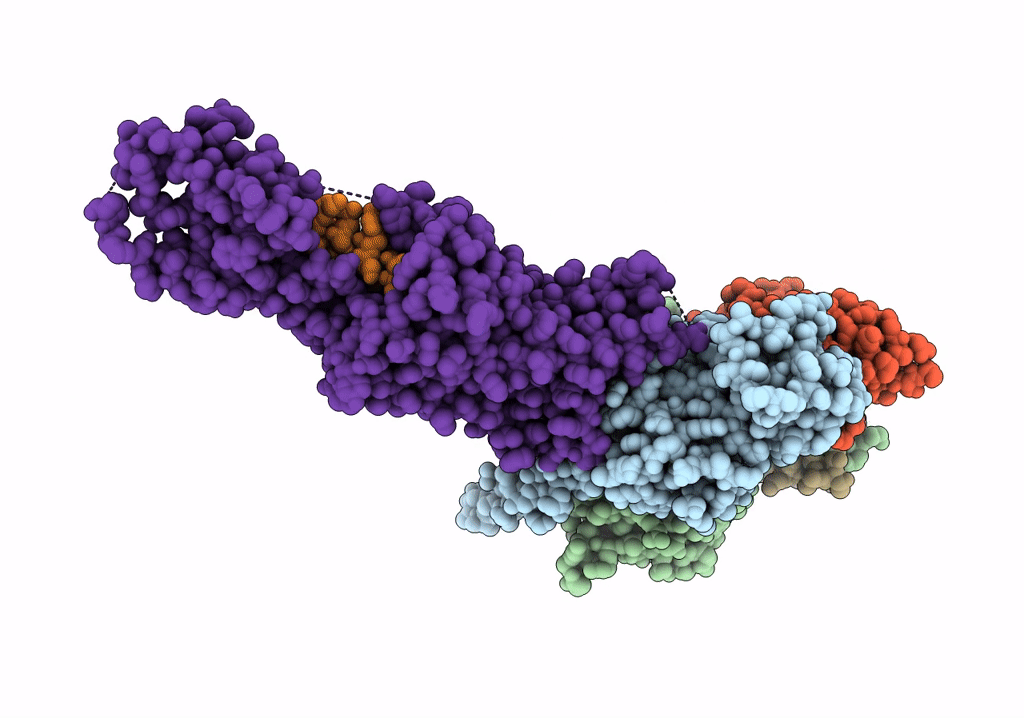 |
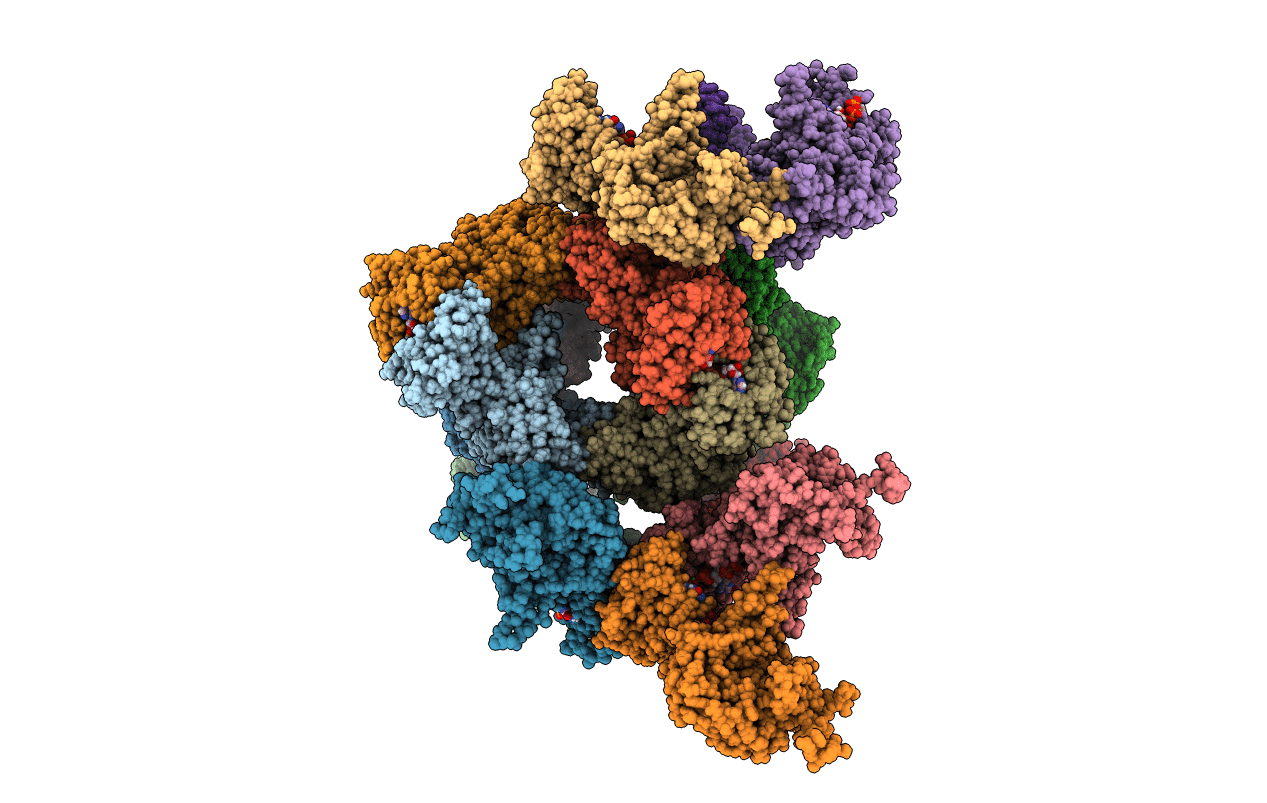
Cryo-Em Structure Of The Glucagon-Like Peptide-1 Receptor In Complex With G Protein, Glp-1 Peptide And A Positive Allosteric Modulator
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2020-07-22 Classification: SIGNALING PROTEIN/MEMBRANE PROTEIN Ligands: QW7 |
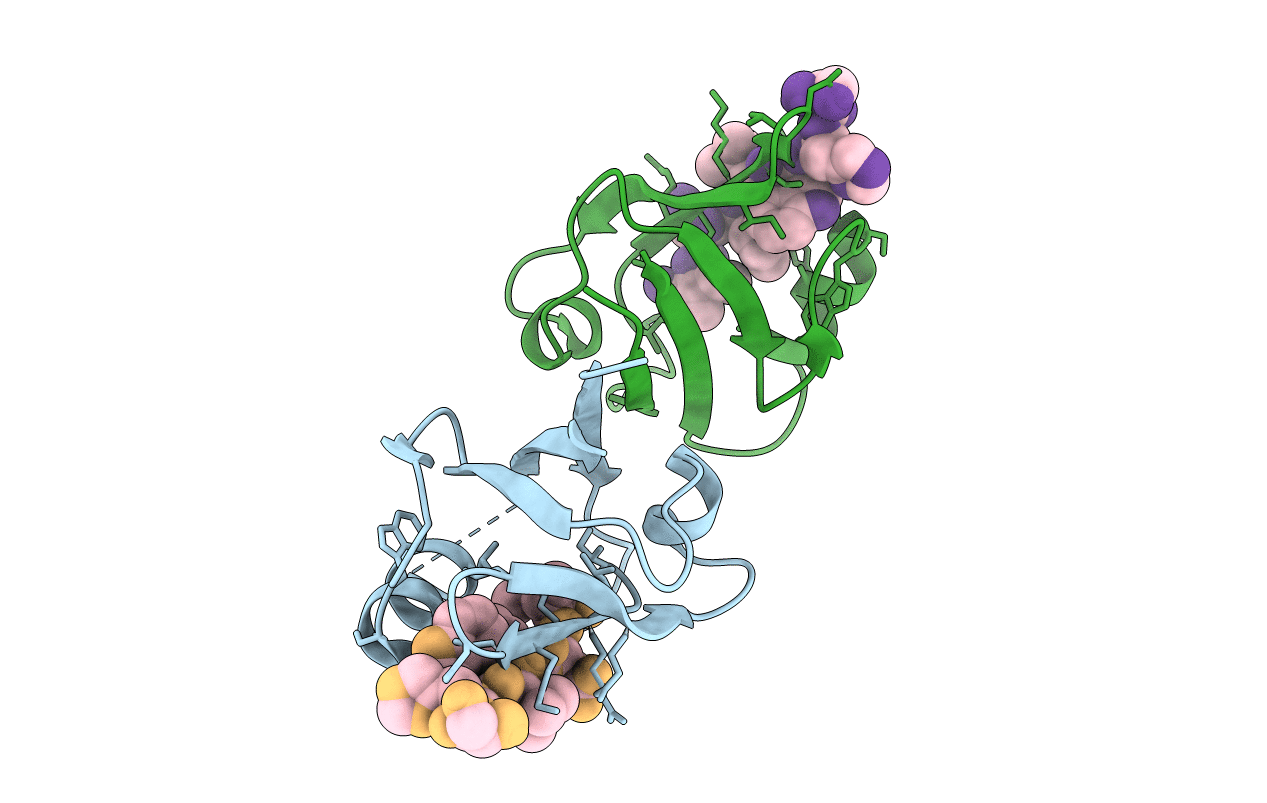 |
Crystal Structure Of The Syntenin Pdz1 Domain In Complex With The Peptide Inhibitor Ksl-128018
Organism: Rattus norvegicus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2019-09-04 Classification: SIGNALING PROTEIN/INHIBITOR |
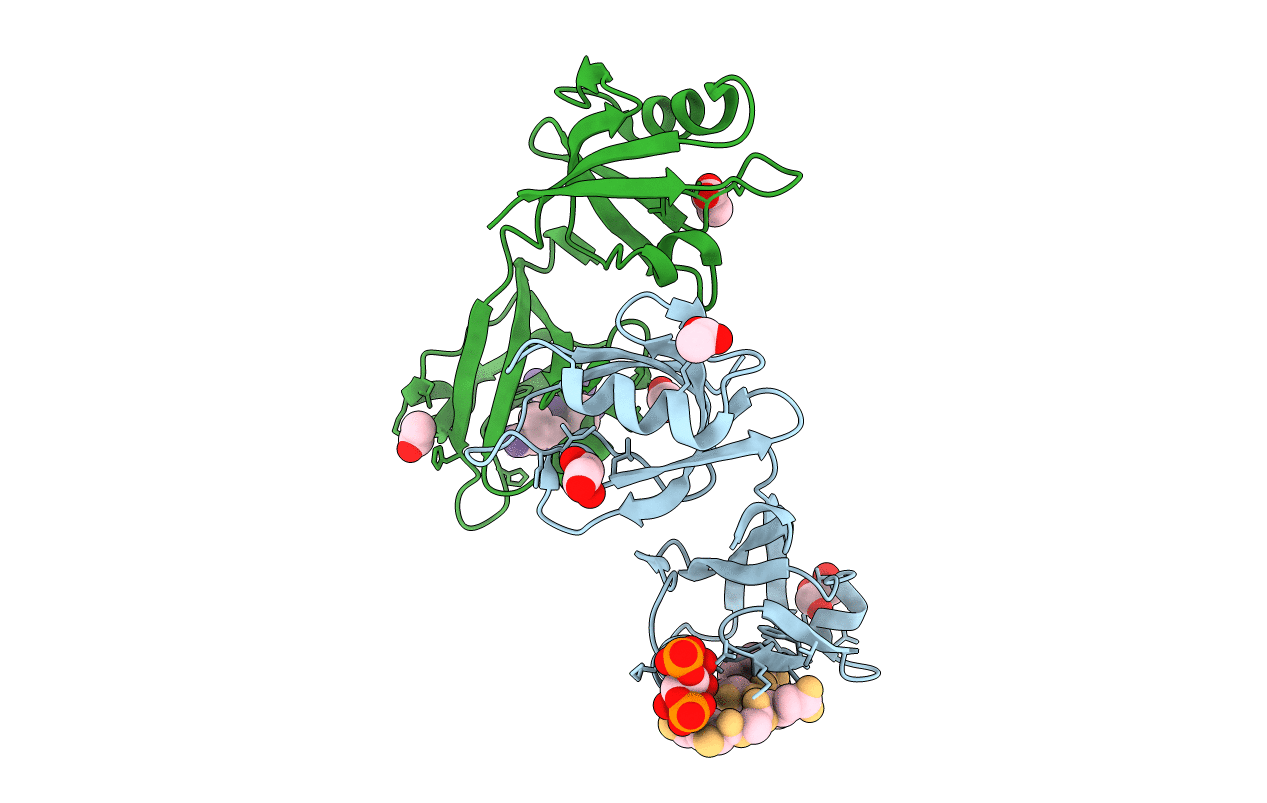 |
Crystal Structure Of The Syntenin Pdz1 And Pdz2 Tandem In Complex With The Frizzled 7 C-Terminal Fragment And Pip2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2016-06-29 Classification: SIGNALING PROTEIN Ligands: ACT, GOL, IP2 |
 |
The Structural Basis Of Dna Binding By The Single-Stranded Dna-Binding Protein From Sulfolobus Solfataricus
Organism: Sulfolobus solfataricus
Method: SOLUTION NMR Release Date: 2014-12-17 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2007-02-27 Classification: TRANSFERASE Ligands: SO4 |
 |
Structure Of The Wild-Type C3Bot1 Exoenzyme (Nicotinamide-Bound State, Crystal Form I)
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2007-02-27 Classification: TRANSFERASE Ligands: NCA, SO4 |
 |
Structure Of The Artt Motif Q212A Mutant C3Bot1 Exoenzyme (Free State, Crystal Form Ii)
Organism: Clostridium botulinum d bacteriophage
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2007-02-27 Classification: TRANSFERASE Ligands: SO4 |
 |
Structure Of The Artt Motif Q212A Mutant C3Bot1 Exoenzyme (Nad-Bound State, Crystal Form I)
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2007-02-27 Classification: TRANSFERASE Ligands: NAD, ADP |
 |
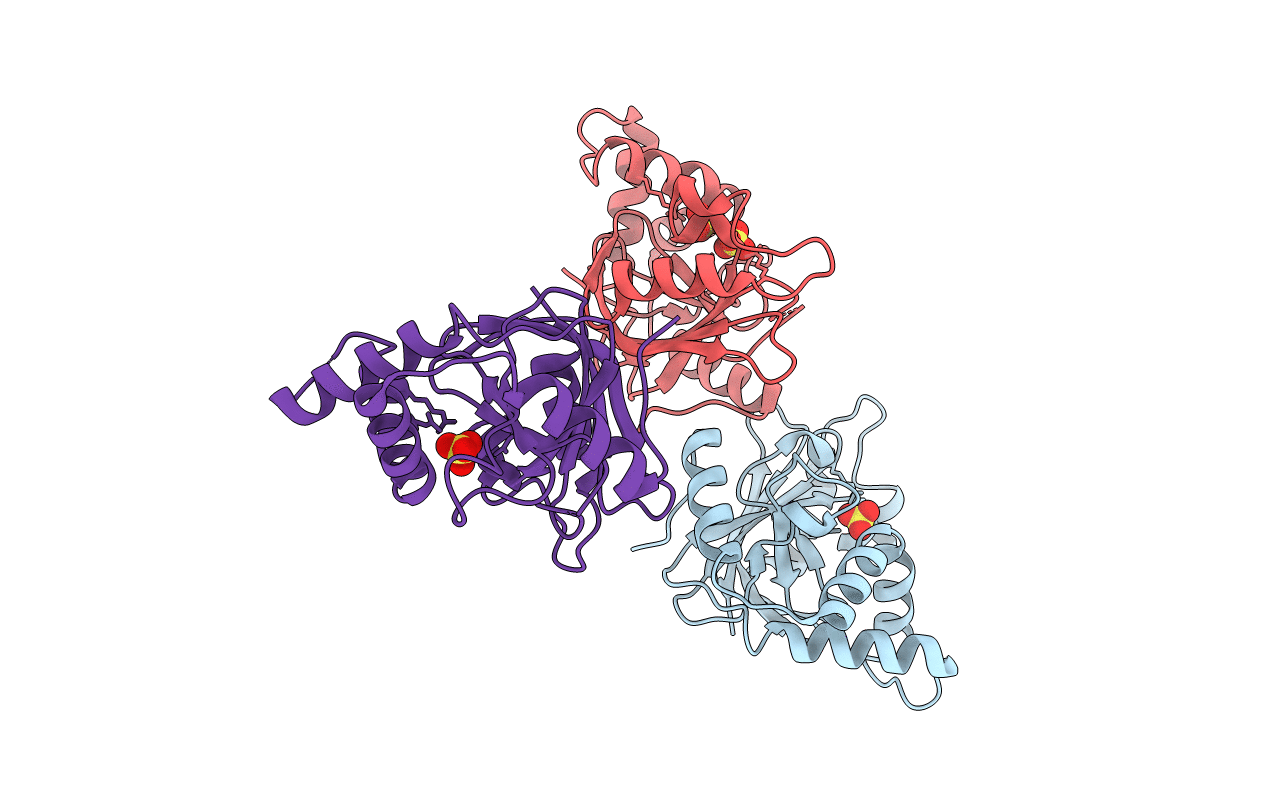
Structure Of The Artt Motif Q212A Mutant C3Bot1 Exoenzyme (Free State, Crystal Form I)
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-02-27 Classification: TRANSFERASE Ligands: SO4 |
 |
Structure Of The Artt Motif E214N Mutant C3Bot1 Exoenzyme (Free State, Crystal Form Iii)
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2007-02-27 Classification: TRANSFERASE Ligands: SO4 |
 |
Structure Of The Artt Motif E214N Mutant C3Bot1 Exoenzyme (Nad-Bound State, Crystal Form Iii)
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-02-27 Classification: TRANSFERASE Ligands: NAD |
 |
Structure Of The Pn Loop Q182A Mutant C3Bot1 Exoenzyme (Free State, Crystal Form I)
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-02-27 Classification: TRANSFERASE Ligands: SO4 |
 |
Structure Of The Pn Loop Q182A Mutant C3Bot1 Exoenzyme (Nad-Bound State, Crystal Form I)
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2007-02-27 Classification: TRANSFERASE Ligands: NAD, SO4 |
 |
General Structure-Based Approach To The Design Of Protein Ligands: Application To The Design Of Kv1.2 Potassium Channel Blockers.
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2006-11-21 Classification: Immune System/DNA |

