Search Count: 17
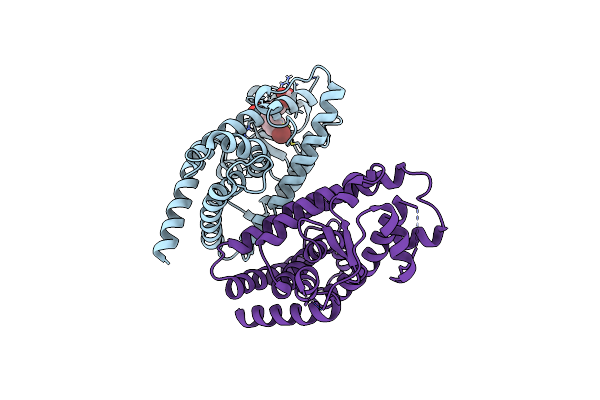 |
X-Ray Crystal Structure Of Ppar Gamma Ligand Binding Domain In Complex With Cz58
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2025-05-14 Classification: TRANSCRIPTION Ligands: A1IYE |
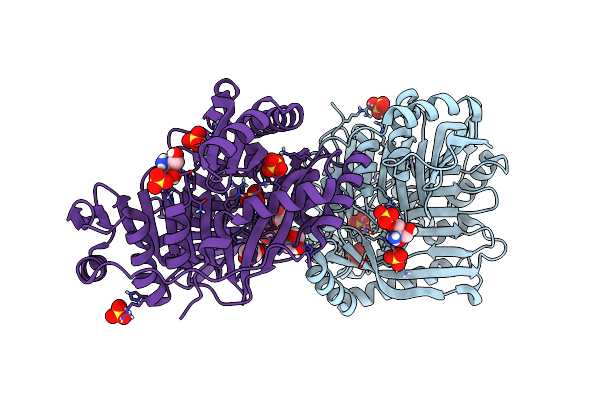 |
Crystallographic Structure Of A Salicylate Synthase From M. Abscessus (Mab-Sas)
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2024-01-31 Classification: BIOSYNTHETIC PROTEIN Ligands: TRS, GOL, SO4, PEG |
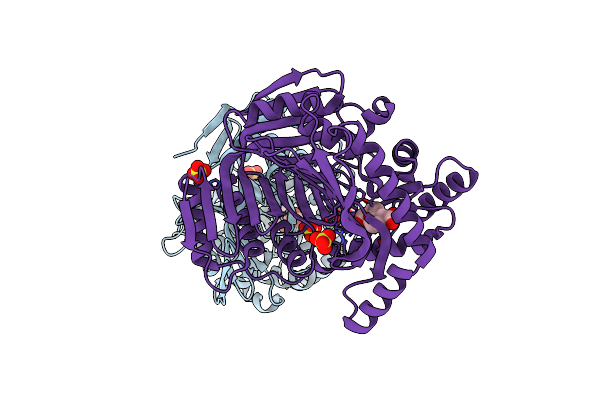 |
M. Tuberculosis Salicylate Synthase Mbti In Complex With 5-(3-Carboxyphenyl)Furan-2-Carboxylic Acid
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2023-11-15 Classification: LYASE Ligands: TXR, SO4, GOL |
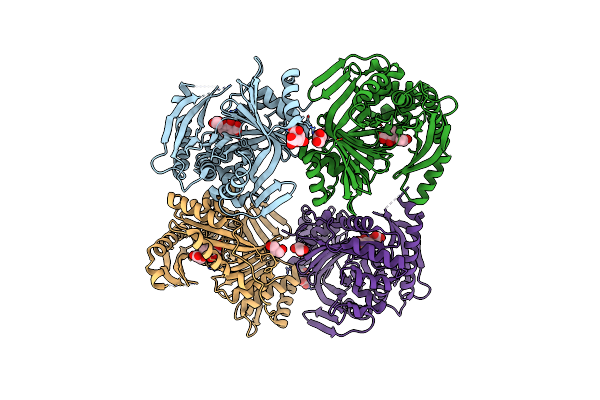 |
M. Tuberculosis Salicylate Synthase Mbti In Complex With Methyl-Amt (New Crystal Form)
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2023-11-15 Classification: LYASE Ligands: 0GA, GOL, FLC, NH4 |
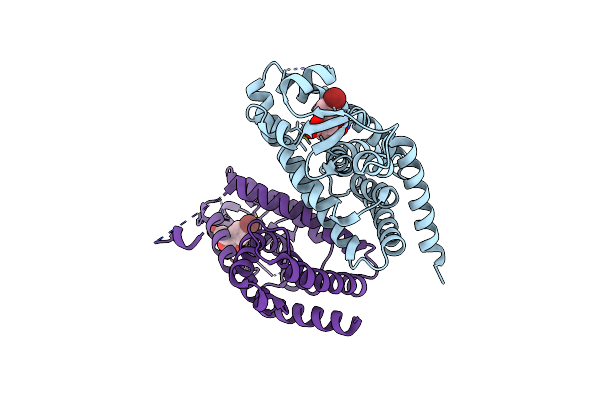 |
X-Ray Crystal Structure Of Ppar Gamma Ligand Binding Domain In Complex With Cz39
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2023-04-26 Classification: TRANSCRIPTION Ligands: LRA |
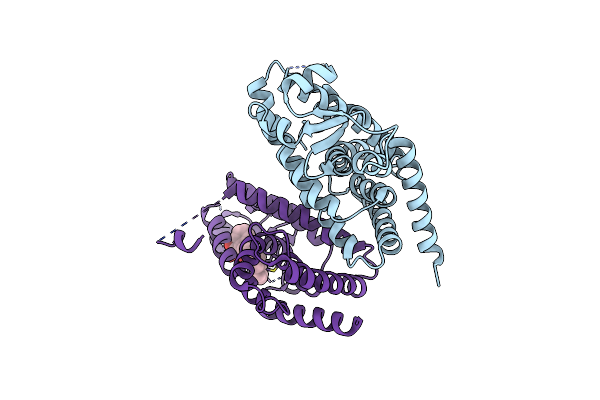 |
X-Ray Crystal Structure Of Ppar Gamma Ligand Binding Domain In Complex With Cz46
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-04-26 Classification: NUCLEAR PROTEIN Ligands: T0C |
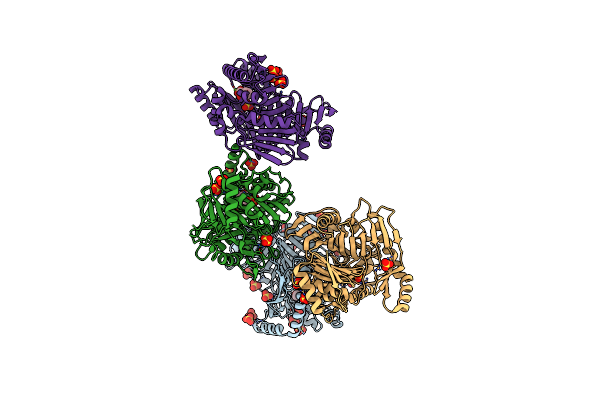 |
M. Tuberculosis Salicylate Synthase Mbti In Complex With Salicylate And Mg2+
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2020-07-15 Classification: LYASE Ligands: SO4, MG, ACT, SAL |
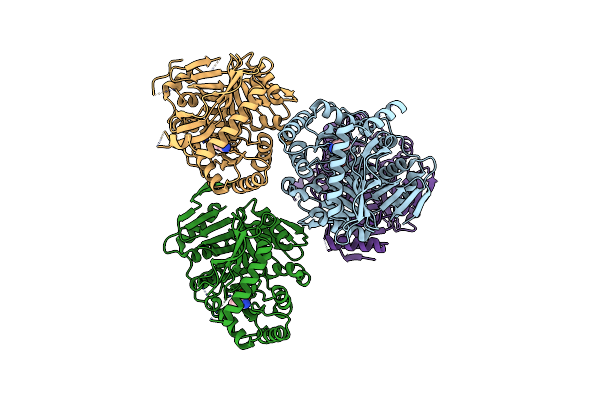 |
M. Tuberculosis Salicylate Synthase Mbti In Complex With 5-(3-Cyanophenyl)Furan-2-Carboxylate
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2020-07-01 Classification: LYASE Ligands: M83, CL |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-07-01 Classification: LYASE Ligands: GOL, PO4, CL, BA |
 |
Organism: Desulfovibrio vulgaris subsp. vulgaris
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-12-21 Classification: ELECTRON TRANSPORT Ligands: FMN |
 |
Low Temperature (100K) Crystal Structure Of Flavodoxin Mutant S64C, Dimer, Semiquinone State
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2004-12-07 Classification: ELECTRON TRANSPORT Ligands: FMN |
 |
Low Temperature (100K) Crystal Structure Of Flavodoxin Mutant S64C, Monomer, Semiquinone State
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2004-12-07 Classification: ELECTRON TRANSPORT Ligands: FMN |
 |
Low Temperature (100K) Crystal Structure Of Flavodoxin Mutant S64C, Homodimer, Oxidised State
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2004-12-07 Classification: ELECTRON TRANSPORT Ligands: FMN |
 |
Organism: desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-11-23 Classification: ELECTRON TRANSPORT Ligands: FMN |
 |
Low Temperature (100K) Crystal Structure Of Flavodoxin D. Vulgaris Wild-Type At 1.35 Angstrom Resolution
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2001-09-05 Classification: ELECTRON TRANSPORT Ligands: FMN |
 |
Low Temperature (100K) Crystal Structure Of Flavodoxin D. Vulgaris S35C Mutant At 1.44 Angstrom Resolution
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2001-09-05 Classification: ELECTRON TRANSPORT Ligands: FMN |
 |
Low Temperature (100K) Crystal Structure Of Flavodoxin D. Vulgaris S64C Mutant, Monomer Oxidised, At 2.4 Angstrom Resolution
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2001-09-05 Classification: ELECTRON TRANSPORT Ligands: FMN |

