Search Count: 18
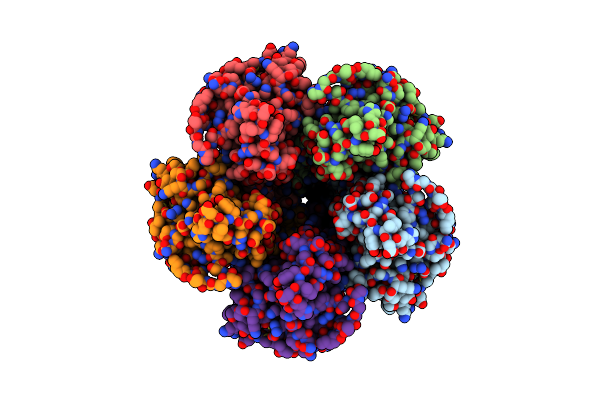 |
Organism: Thermochaetoides thermophila
Method: ELECTRON MICROSCOPY Release Date: 2024-11-27 Classification: MEMBRANE PROTEIN Ligands: MG |
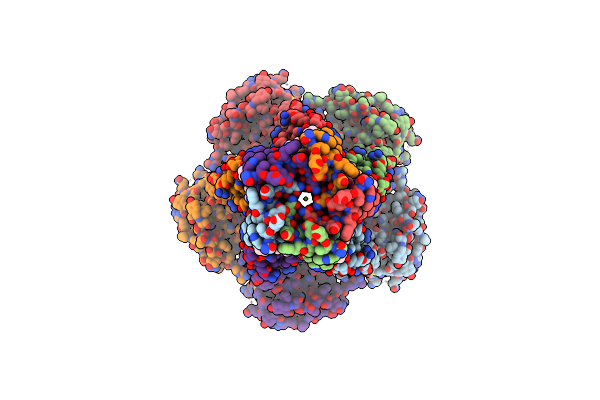 |
Organism: Thermochaetoides thermophila
Method: ELECTRON MICROSCOPY Release Date: 2024-11-27 Classification: MEMBRANE PROTEIN Ligands: LOP, MG |
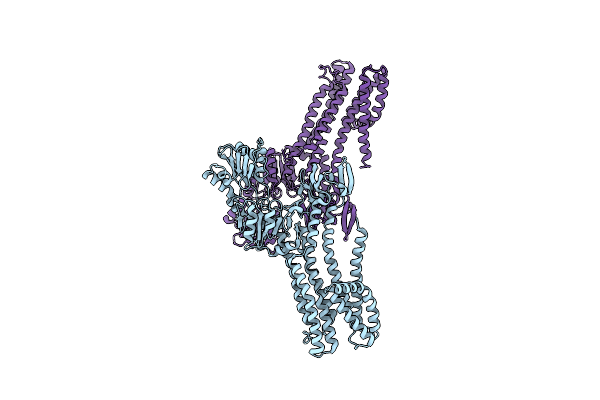 |
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:4.01 Å Release Date: 2022-09-14 Classification: MEMBRANE PROTEIN |
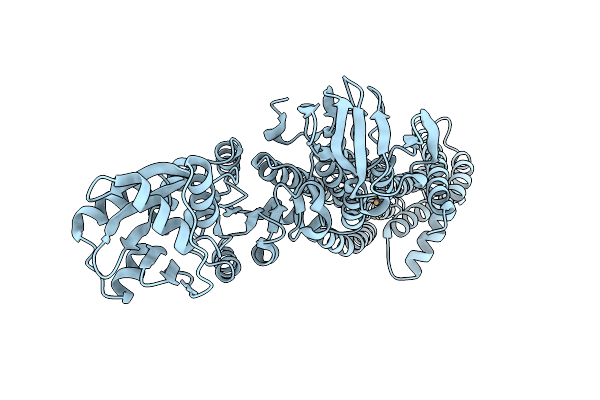 |
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:3.31 Å Release Date: 2022-09-14 Classification: MEMBRANE PROTEIN Ligands: CU |
 |
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-09-14 Classification: MEMBRANE PROTEIN Ligands: K, MG |
 |
Organism: Thermochaetoides thermophila dsm 1495
Method: ELECTRON MICROSCOPY Release Date: 2022-08-10 Classification: MEMBRANE PROTEIN |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: ELECTRON MICROSCOPY Release Date: 2022-08-10 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
 |
Structure Of Ctatm1 In The Inward-Open With Glutathione-Complexed [2Fe-2S] Cluster Bound
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: ELECTRON MICROSCOPY Resolution:3.00 Å Release Date: 2022-08-10 Classification: MEMBRANE PROTEIN |
 |
Structure Of Ctatm1 In The Inward-Facing Partially Occluded With Cargo Bound
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: ELECTRON MICROSCOPY Resolution:3.20 Å Release Date: 2022-08-10 Classification: MEMBRANE PROTEIN Ligands: GSH |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: ELECTRON MICROSCOPY Release Date: 2022-08-10 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-07-06 Classification: TRANSPORT PROTEIN Ligands: C8E, LDA, SCN |
 |
Organism: Sulfitobacter sp. (strain nas-14.1)
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2022-03-23 Classification: MEMBRANE PROTEIN Ligands: ALF, MG |
 |
Organism: Sulfitobacter sp. (strain nas-14.1)
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2022-03-23 Classification: MEMBRANE PROTEIN Ligands: BEF, MG |
 |
Organism: Mycolicibacterium thermoresistibile, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-04-01 Classification: MEMBRANE PROTEIN Ligands: NI, DMU |
 |
Organism: Mycolicibacterium thermoresistibile
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-04-01 Classification: MEMBRANE PROTEIN Ligands: FAD |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2017-06-28 Classification: TRANSCRIPTION |
 |
Organism: Shigella sonnei
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2014-08-13 Classification: HYDROLASE Ligands: MG, BEF |
 |
Organism: Shigella sonnei
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2014-08-13 Classification: HYDROLASE Ligands: ALF, MG |

