Search Count: 29
 |
Structure Of The 50S Ribosomal Subunit From The Antibiotic-Producing Bacterium Streptomyces Fradiae
Organism: Streptomyces fradiae atcc 10745 = dsm 40063
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: RIBOSOME |
 |
Organism: Psychrobacter urativorans
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: RIBOSOME |
 |
Organism: Methanosarcina mazei
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: RNA |
 |
Cryo-Em Structure Of P. Urativorans 70S Ribosome In Complex With Hibernation Factors Balon And Raia (Structure 1).
Organism: Psychrobacter urativorans
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: RIBOSOME Ligands: MG |
 |
Cryo-Em Structure Of P. Urativorans 70S Ribosome In Complex With Hibernation Factor Balon, Mrna And P-Site Trna (Structure 2).
Organism: Psychrobacter urativorans
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: RIBOSOME |
 |
Cryo-Em Structure Of P. Urativorans 70S Ribosome In Complex With Hibernation Factor Balon And Ef-Tu(Gdp) (Structure 3).
Organism: Psychrobacter urativorans
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: RIBOSOME Ligands: GDP, MG |
 |
Cryo-Em Structure Of The Mycobacterium Smegmatis 70S Ribosome In Complex With Hibernation Factor Msmeg1130 (Balon) (Structure 4)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: RIBOSOME Ligands: ZN |
 |
Cryo-Em Structure Of The Mycobacterium Smegmatis 70S Ribosome In Complex With Hibernation Factor Rv2629 (Balon) (Structure 5)
Organism: Mycobacterium tuberculosis h37rv, Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: RIBOSOME Ligands: ZN |
 |
Cryo-Em Structure Of The Mycobacterium Smegmatis 70S Ribosome In Complex With Hibernation Factor Msmeg1130 (Balon) And Msmegef-Tu(Gdp) (Composite Structure 6)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: RIBOSOME Ligands: GDP, ZN |
 |
Organism: Candidatus methanomethylophilus alvus
Method: ELECTRON MICROSCOPY Release Date: 2024-01-10 Classification: RNA |
 |
Organism: Encephalitozoon cuniculi gb-m1
Method: ELECTRON MICROSCOPY Release Date: 2022-02-09 Classification: RIBOSOME Ligands: ZN, AMP, SPD |
 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With A Short Substrate Mimic Cc-Pmn And Bound To Mrna And P-Site Trna At 3.7A Resolution
Organism: Escherichia coli, Enterobacteria phage t4, Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2018-12-12 Classification: RIBOSOME Ligands: MG, ZN, SF4 |
 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With A Short Substrate Mimic Acca-Dphe And Bound To Mrna And P-Site Trna At 3.7A Resolution
Organism: Escherichia coli, Enterobacteria phage t4, Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2018-12-12 Classification: RIBOSOME Ligands: MG, ZN, SF4 |
 |
Cryo-Em Reconstruction Of Yeast 80S Ribosome In Complex With Mrna, Trna And Eef2 (Gmppcp/Sordarin)
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2018-07-11 Classification: RIBOSOME Ligands: ZN, SO1, GCP |
 |
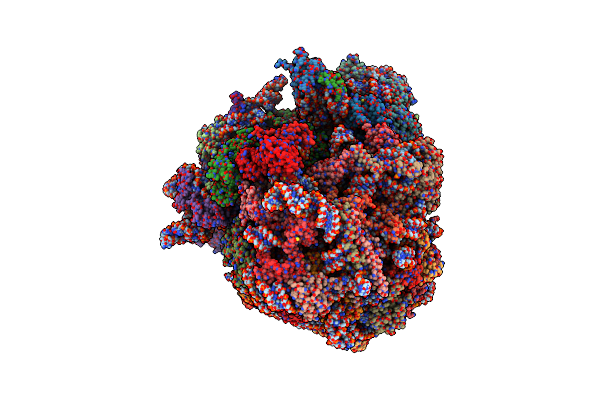
Cryo-Em Reconstruction Of Yeast 80S Ribosome In Complex With Mrna, Trna And Eef2 (Gdp+Alf4/Sordarin)
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2018-07-11 Classification: RIBOSOME Ligands: ZN, SO1, GDP, ALF |
 |
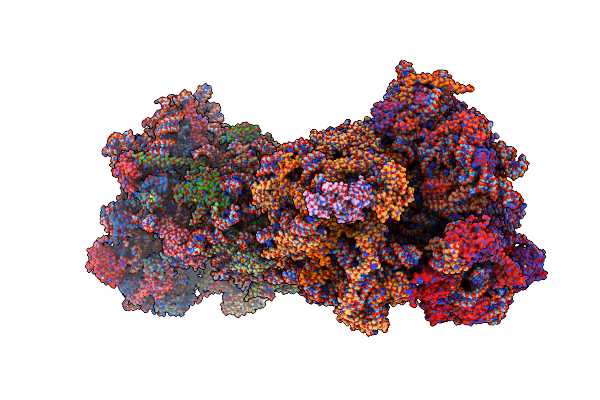
Cryo-Em Recosntruction Of Yeast 80S Ribosome In Complex With Mrna, Trna And Eef2 (Gmppcp)
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2018-07-11 Classification: RIBOSOME Ligands: ZN, GCP |
 |
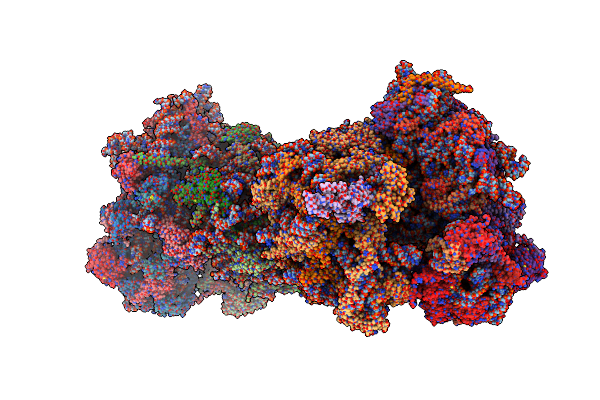
Coping With Proline Stalling: Structural Basis Of Hypusine-Induced Protein Synthesis By The Eukaryotic Ribosome
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2017-01-25 Classification: RIBOSOME Ligands: MG, OHX, ZN, SPS, PRO |
 |
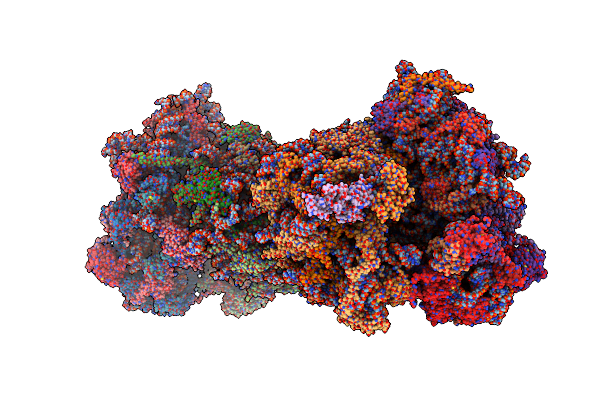
Crystal Structure Of The S.Cerevisiae 80S Ribosome In Complex With The A-Site Bound Aminoacyl-Trna Analog Acca-Pro
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2017-01-18 Classification: RIBOSOME Ligands: OHX, MG, ZN, PHE, LEU, SPS, 8AN |
 |
Complex Of Yeast 80S Ribosome With Hypusine-Containing/Non-Modified Eif5A And/Or A Peptidyl-Trna Analog
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2016-12-14 Classification: RIBOSOME Ligands: ZN, SPS, MG, OHX |
 |
Complex Of Yeast 80S Ribosome With Hypusine-Containing/Non-Modified Eif5A And/Or A Peptidyl-Trna Analog
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2016-12-14 Classification: RIBOSOME Ligands: ZN, SPS, MG |

