Search Count: 15
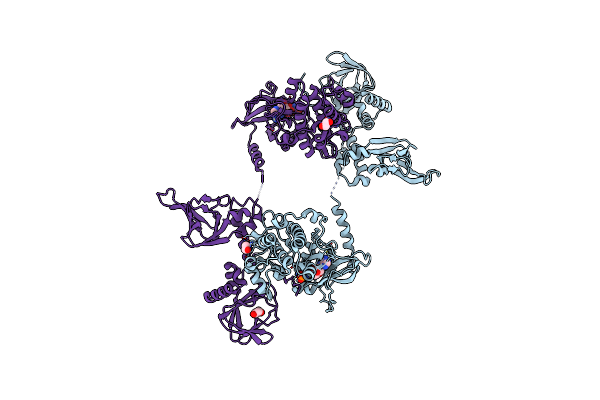 |
Crystal Structure Of Human Protein Kinase G (Pkg) R-C Complex In Inhibited State
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2022-08-24 Classification: TRANSFERASE/INHIBITOR Ligands: MN, ANP, EDO |
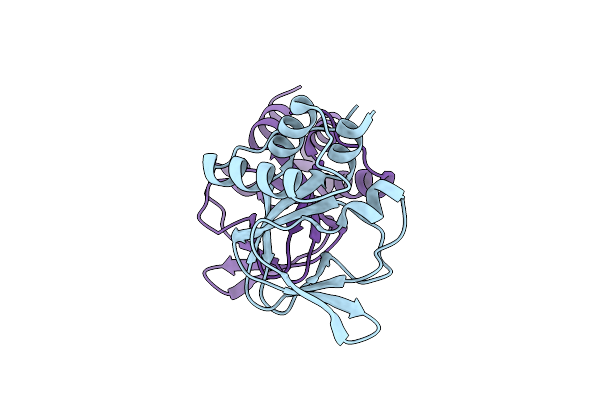 |
Crystal Structure Of Cgmp Dependent Protein Kinase I Alpha (Pkg I Alpha)Cnb-A Domain With R177Q Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2022-05-11 Classification: SIGNALING PROTEIN |
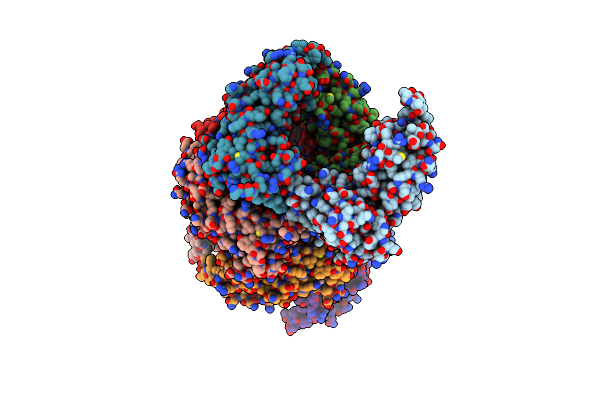 |
Crystal Structure Of A211D Mutant Of Protein Kinase A Ria Subunit, A Carney Complex Mutation
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:4.16 Å Release Date: 2021-05-26 Classification: TRANSFERASE Ligands: CMP |
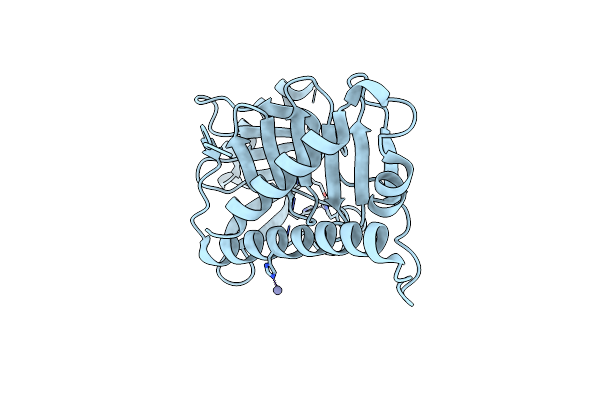 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-02-19 Classification: CELL CYCLE, Transferase Ligands: ZN |
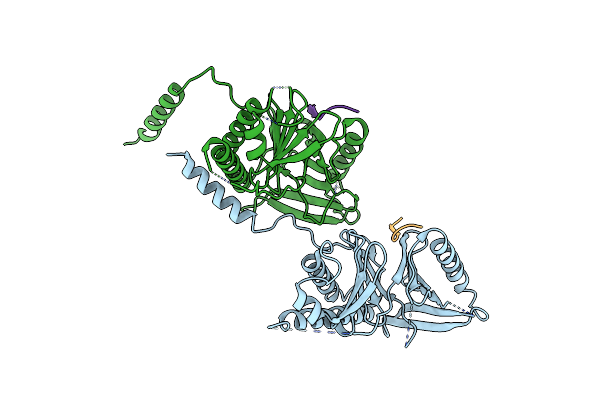 |
Crystal Structure Of Budding Yeast Cdc5 Polo-Box Domain In Complex With Spc72 Phosphopeptide.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-02-19 Classification: CELL CYCLE, Transferase Ligands: CL |
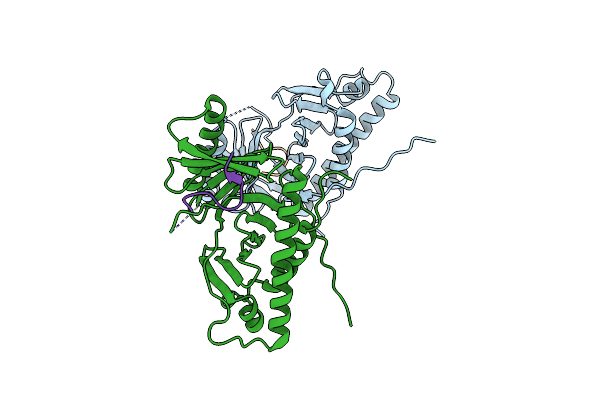 |
Crystal Structure Of Budding Yeast Cdc5 Polo-Box Domain In Complex With The Dbf4 Polo-Interacting Region.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2020-02-19 Classification: CELL CYCLE, Transferase |
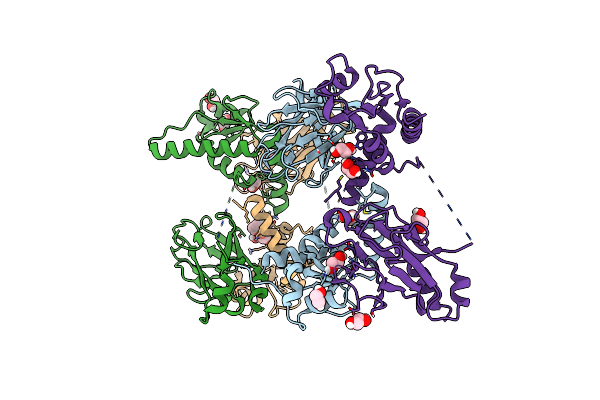 |
Organism: Saccharomyces cerevisiae, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2017-03-15 Classification: CELL CYCLE Ligands: EDO |
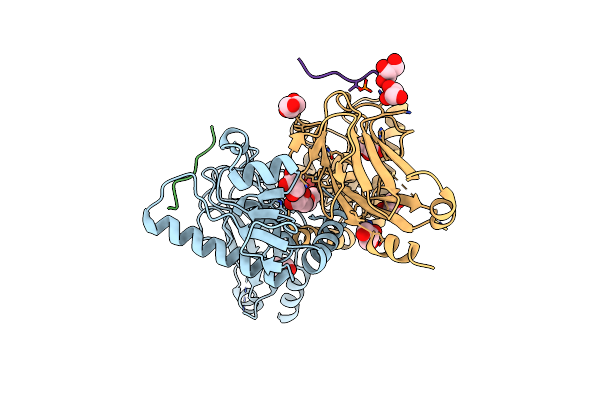 |
Structure Of The Fha1 Domain Of Rad53 Bound Simultaneously To The Brct Domain Of Dbf4 And A Phosphopeptide.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-10-12 Classification: CELL CYCLE Ligands: GOL |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2014-06-04 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-01-15 Classification: SIGNALING PROTEIN Ligands: IOD, PCG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2014-01-15 Classification: SIGNALING PROTEIN Ligands: GLY |
 |
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2000-01-01 Classification: TRANSCRIPTION/DNA |
 |
Organism: Enterobacteria phage p22
Method: SOLUTION NMR Release Date: 1999-11-03 Classification: TRANSLATION/REGULATION |
 |
Nmr Study Of The Backbone Conformational Equilibria Of Sandostatin, Minimized Average Beta-Sheet Structure
|
 |
Nmr Study Of The Backbone Conformational Equilibria Of Sandostatin, Two Representative Minimum Energy Partially Helical Structures
|

