Search Count: 18
 |
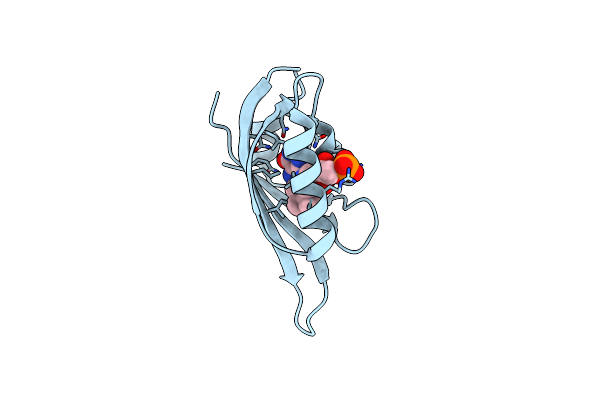
Structure Of Meiothermus Ruber Mrub_1259 Lov Domain With N- And C-Terminal Alpha Helices (Mrlove)
Organism: Meiothermus ruber dsm 1279
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: FLUORESCENT PROTEIN Ligands: FMN |
 |
Organism: Meiothermus ruber dsm 1279
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: FLUORESCENT PROTEIN Ligands: FMN |
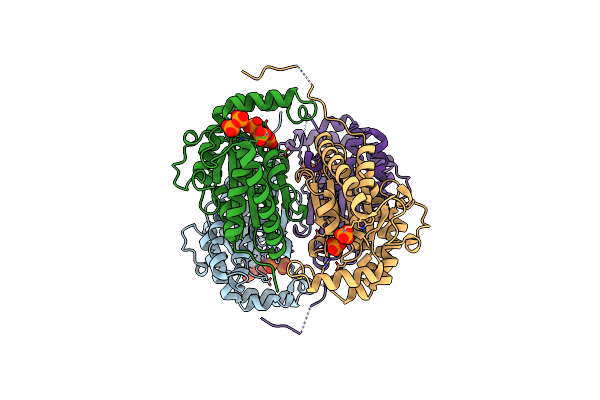 |
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Release Date: 2018-03-21 Classification: TRANSFERASE Ligands: PO4, DPO |
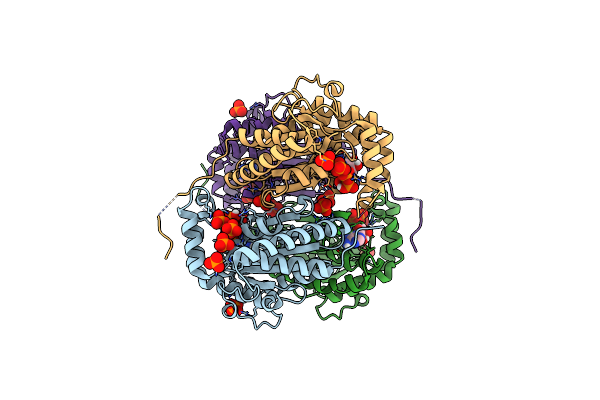 |
Structure Of Polyphosphate Kinase From Meiothermus Ruber N121D Bound To Atp
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-03-21 Classification: TRANSFERASE Ligands: ATP, PO4 |
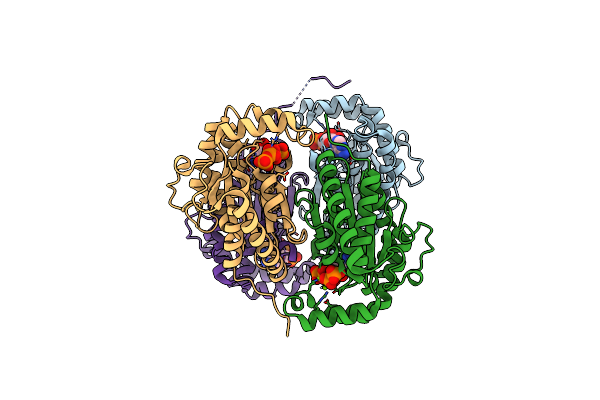 |
Crystal Structure Of Polyphosphate Kinase From Meiothermus Ruber Bound To Adp And Ppi
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2017-12-20 Classification: TRANSFERASE Ligands: ADP, PPV, MG |
 |
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-06-21 Classification: TRANSFERASE Ligands: PO4, SO4 |
 |
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2017-06-21 Classification: TRANSFERASE Ligands: AMP, PO4, SO4, MG |
 |
Crystal Structure Of Polyphosphate Kinase From Meiothermus Ruber Bound To Atp
Organism: Meiothermus ruber
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2017-06-21 Classification: TRANSFERASE Ligands: ATP, MG, SO4, GOL |
 |
Crystal Structure Of Polyphosphate Kinase From Meiothermus Ruber Bound To Adp
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-06-21 Classification: TRANSFERASE Ligands: ADP, PO4, MG, GOL, CL |
 |
Organism: Meiothermus ruber
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-04-30 Classification: RNA BINDING PROTEIN Ligands: HG |
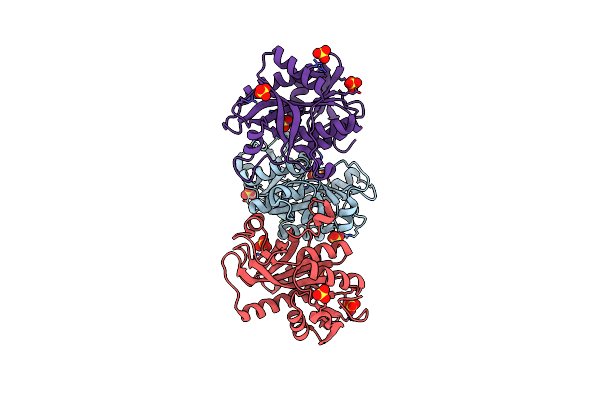 |
Crystal Structure Of Purine Nucleoside Phosphorylase From Meiothermus Ruber Dsm 1279 Complexed With Sulfate.
Organism: Meiothermus ruber
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2013-10-09 Classification: TRANSFERASE Ligands: MG, SO4 |
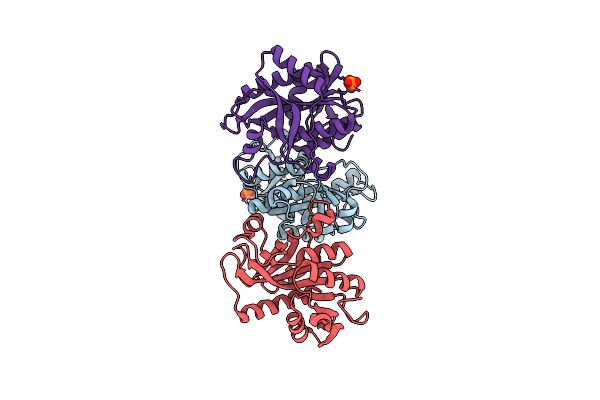 |
Crystal Structure Of Purine Nucleoside Phosphorylase From Meiothermus Ruber Dsm 1279, Nysgrc Target 029804.
Organism: Meiothermus ruber dsm 1279
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-08-28 Classification: TRANSFERASE Ligands: PO4, MG |
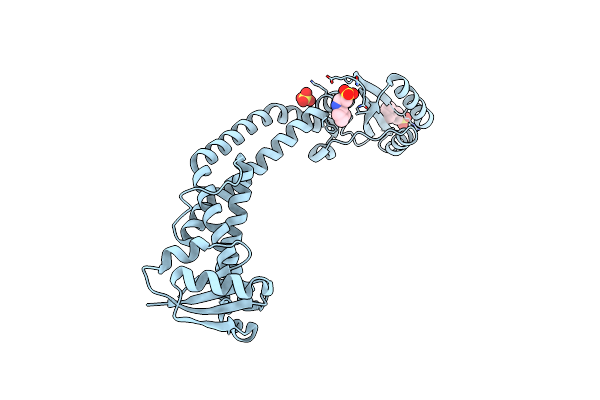 |
Crystal Structure Of The Cytoplasmic N-Terminal Domain Of Subunit I, Homolog Of Subunit A, Of V-Atpase
Organism: Meiothermus ruber
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2011-08-10 Classification: PROTON TRANSPORT Ligands: NHE, SO4 |
 |
NA
Organism: Meiothermus ruber (strain ATCC 35948 / DSM 1279 / VKM B-1258 / 21)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
|
 |
NA
|
 |
NA
|
 |
NA
|

