Search Count: 131
 |
Crystal Structure Of The Poly(Hexamethylene Adipamide) (Nylon66) Hydrolase Nyl50 In Its Apo Form At Room Temperature
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: HYDROLASE |
 |
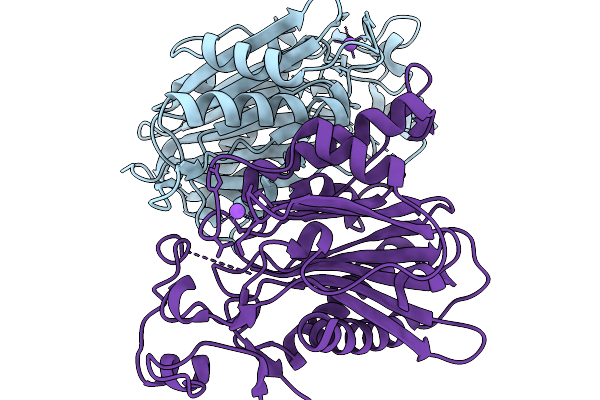
Joint Xray/Neutron Structure Of Macrophage Migration Inhibitory Factor (Mif) Bound To 4-Hydroxyphenylpyruvate At Room Temperature
Organism: Homo sapiens
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-08-27 Classification: CYTOKINE Ligands: ENO, IPA, EN1 |
 |
Joint Neutron And X-Ray Structure Of Alginate Lyase Psalg7C Soaked With Pentamannuronic Acid
Organism: Paradendryphiella salina
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-07-30 Classification: LYASE |
 |
Organism: Paradendryphiella salina
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-07-30 Classification: LYASE |
 |
Crystal Structure Of Paradendryphiella Salina Pl7C Alginate Lyase Mutant H124N Soaked With With Penta-Mannuronic Acid
Organism: Paradendryphiella salina
Method: X-RAY DIFFRACTION Release Date: 2025-02-05 Classification: LYASE |
 |
X-Ray Crystallographic Structure Of The Poly(Hexamethylene Adipamide) (Nylon66) Hydrolase Nyl50 At Room Temperature
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-01-29 Classification: HYDROLASE Ligands: EDO, NA, PGE |
 |
X-Ray Crystallographic Structure Of The Poly(Hexamethylene Adipamide) (Nylon66) Hydrolase Nyl50 At Room Temperature Bound To Tetraethylene Glycol
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-01-29 Classification: HYDROLASE Ligands: EDO, NA, PG4 |
 |
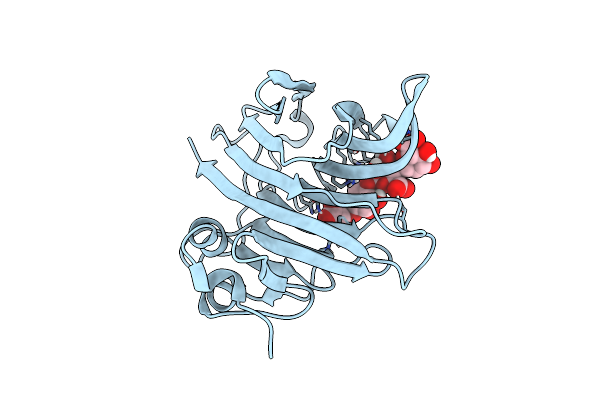
Crystal Structure Of Paradendryphiella Salina Pl7C Alginate Lyase Mutant H124 Soaked With Hexa-Mannuronic Acid
Organism: Paradendryphiella salina
Method: X-RAY DIFFRACTION Release Date: 2024-10-02 Classification: LYASE |
 |
Crystal Structure Of Paradendryphiella Salina Pl7C Alginate Lyase Mutant H124 Soaked With With Tetra-Mannuronic Acid
Organism: Paradendryphiella salina
Method: X-RAY DIFFRACTION Release Date: 2024-09-11 Classification: LYASE |
 |
X-Ray Structure Of Macrophage Migration Inhibitory Factor (Mif) Covalently Bound To 4-Hydroxyphenylpyruvate (Hpp)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-08-28 Classification: CYTOKINE Ligands: ENO, IPA |
 |
Crystal Structure Of Paradendryphiella Salina Pl7C Alginate Lyase Mutant H110N In Complex With Tri-Mannuronic Acid
Organism: Paradendryphiella salina
Method: X-RAY DIFFRACTION Release Date: 2024-01-17 Classification: LYASE |
 |
Crystal Structure Of Paradendryphiella Salina Pl7C Alginate Lyase Mutant H124
Organism: Paradendryphiella salina
Method: X-RAY DIFFRACTION Release Date: 2024-01-17 Classification: LYASE Ligands: PEG, NA |
 |
Crystal Structure Of Paradendryphiella Salina Pl7C Alginate Lyase Mutant Y220F
Organism: Paradendryphiella salina
Method: X-RAY DIFFRACTION Release Date: 2023-12-27 Classification: LYASE Ligands: ACT, SO4 |
 |
Crystal Structure Of Paradendryphiella Salina Pl7C Alginate Lyase Mutant Y220F In Complex With Penta-Mannuronic Acid
Organism: Paradendryphiella salina
Method: X-RAY DIFFRACTION Release Date: 2023-12-20 Classification: LYASE Ligands: SO4 |
 |
Crystal Structure Of Paradendryphiella Salina Pl7C Alginate Lyase Mutant Y220F In Complex With Hexa-Mannuronic Acid
Organism: Paradendryphiella salina
Method: X-RAY DIFFRACTION Release Date: 2023-11-15 Classification: LYASE Ligands: SO4 |
 |
Room Temperature Neutron Structure Of A Fluorescent Ag8 Cluster Templated By A Multistranded Dna Scaffold
Organism: Synthetic construct
Method: NEUTRON DIFFRACTION Resolution:2.10 Å Release Date: 2023-08-02 Classification: DNA Ligands: AG |
 |
Crystal Structure Of Paradendryphiella Salina Pl7C Alginate Lyase In Complex With Pentamannuronic Acid
Organism: Paradendryphiella salina
Method: X-RAY DIFFRACTION Release Date: 2023-07-12 Classification: LYASE Ligands: CL |
 |
Crystal Structure Of Paradendryphiella Salina Pl7C Alginate Lyase Soaked With Hexamannuronic Acid
Organism: Paradendryphiella salina
Method: X-RAY DIFFRACTION Release Date: 2023-07-12 Classification: LYASE |
 |
Crystal Structure Of Paradendryphiella Salina Pl7C Alginate Lyase Soaked With Tetramannuronic Acid
Organism: Paradendryphiella salina
Method: X-RAY DIFFRACTION Release Date: 2023-07-12 Classification: LYASE Ligands: CL |
 |
Crystal Structure Of Paradendryphiella Salina Pl7C Alginate Lyase Soaked With Dimannuronic Acid
Organism: Paradendryphiella salina
Method: X-RAY DIFFRACTION Release Date: 2023-07-12 Classification: LYASE |

