Search Count: 33
 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: MEMBRANE PROTEIN Ligands: HEM, HEC, FES |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-08-23 Classification: HYDROLASE Ligands: NUX |
 |
Organism: Acinetobacter baumannii (strain atcc 17978 / cip 53.77 / lmg 1025 / ncdc kc755 / 5377)
Method: ELECTRON MICROSCOPY Release Date: 2022-02-02 Classification: MEMBRANE PROTEIN Ligands: ATP, MG, ADP, PO4 |
 |
Organism: Acinetobacter baumannii atcc 17978
Method: ELECTRON MICROSCOPY Release Date: 2022-02-02 Classification: MEMBRANE PROTEIN Ligands: ATP, MG, ADP |
 |
Organism: Acinetobacter baumannii (strain atcc 17978 / cip 53.77 / lmg 1025 / ncdc kc755 / 5377)
Method: ELECTRON MICROSCOPY Release Date: 2022-02-02 Classification: MEMBRANE PROTEIN Ligands: ATP, MG, ADP |
 |
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Resolution:3.15 Å Release Date: 2018-05-23 Classification: MEMBRANE PROTEIN Ligands: ATP, MG, ADP |
 |
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Release Date: 2018-05-23 Classification: MEMBRANE PROTEIN Ligands: MG, ATP, ADP |
 |
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Release Date: 2018-05-23 Classification: MEMBRANE PROTEIN Ligands: ATP, MG, ADP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2018-01-10 Classification: TRANSFERASE, HYDROLASE Ligands: 8UM, AMZ, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2018-01-10 Classification: TRANSFERASE, HYDROLASE Ligands: AMZ, MG, 8US |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2017-01-11 Classification: HORMONE Ligands: IPH, ZN, CL, NA |
 |
Organism: Yarrowia lipolytica
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2016-06-29 Classification: HYDROLASE Ligands: ATP, MG, ADP |
 |
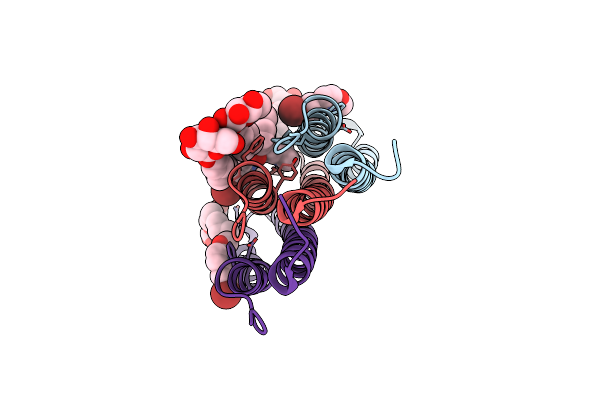
Crystal Structure Of A Mycobacterial Atp Synthase Rotor Ring In Complex With Iodo-Bedaquiline
Organism: Mycobacterium phlei
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-01-20 Classification: HYDROLASE Ligands: IBQ, TAM |
 |
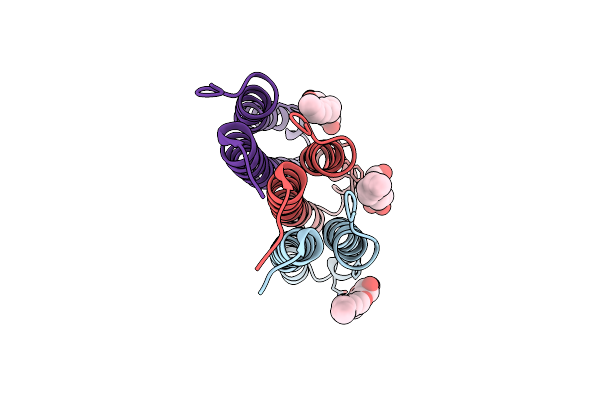
Crystal Structure Of A Mycobacterial Atp Synthase Rotor Ring In Complex With Bedaquiline
Organism: Mycobacterium phlei
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-05-20 Classification: HYDROLASE Ligands: BQ1, BOG |
 |
Organism: Mycobacterium phlei
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2015-05-20 Classification: HYDROLASE Ligands: BOG |
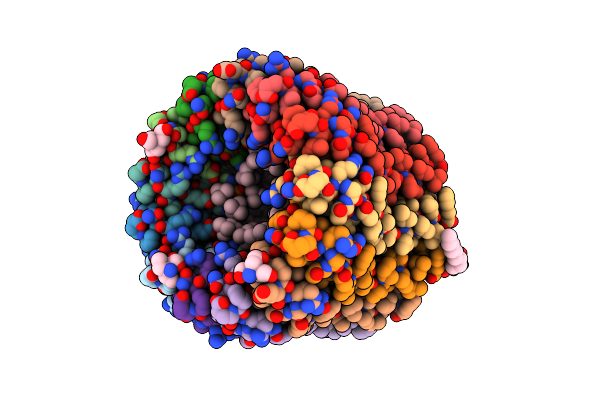 |
The C-Ring Ion Binding Site Of The Atp Synthase From Bacillus Pseudofirmus Of4 Is Adapted To Alkaliphilic Cell Physiology
Organism: Bacillus pseudofirmus of4
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-05-28 Classification: TRANSFERASE Ligands: DPV, TAM, LMT |
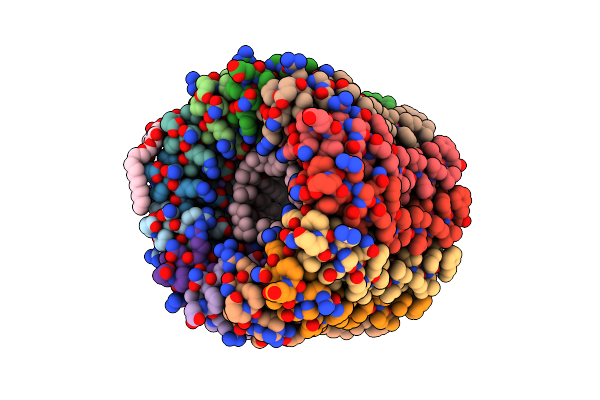 |
The C-Ring Ion Binding Site Of The Atp Synthase From Bacillus Pseudofirmus Of4 Is Adapted To Alkaliphilic Cell Physiology
Organism: Bacillus pseudofirmus of4
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2014-04-23 Classification: TRANSFERASE Ligands: DPV, NA, LMT |
 |
Crystal Structure Of The F-Type Atp Synthase C-Ring From Acetobacterium Woodii.
Organism: Acetobacterium woodii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-03-26 Classification: HYDROLASE |
 |
Organism: Fusobacterium nucleatum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-05-29 Classification: MEMBRANE PROTEIN |
 |
Organism: Fusobacterium nucleatum
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2013-05-29 Classification: MEMBRANE PROTEIN |

