Search Count: 16
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2024-02-07 Classification: APOPTOSIS Ligands: SM, NAG |
 |
Nanodisc Reconstituted Msba In Complex With Nanobodies, Spin-Labeled At Position A60C
Organism: Escherichia coli k-12, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: MEMBRANE PROTEIN Ligands: EIW, 88T, GD |
 |
Amp-Pnp Bound Nanodisc Reconstituted Msba With Nanobodies, Spin-Labeled At Position A60C
Organism: Escherichia coli k-12, Vicugna pacos
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2022-08-24 Classification: MEMBRANE PROTEIN Ligands: ANP, MG, LMT, 3PE, 88T, GD |
 |
Amp-Pnp Bound Nanodisc Reconstituted Msba With Nanobodies, Spin-Labeled At Position T68C
Organism: Escherichia coli (strain k12), Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: MEMBRANE PROTEIN Ligands: ANP, MG, LMT, 88T, GD |
 |
Nanodisc Reconstituted Msba In Complex With Nanobodies, Spin-Labeled At Position T68C
Organism: Escherichia coli, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: MEMBRANE PROTEIN Ligands: EIW, 88T, GD |
 |
Organism: Enterococcus faecalis, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2022-05-18 Classification: MEMBRANE PROTEIN |
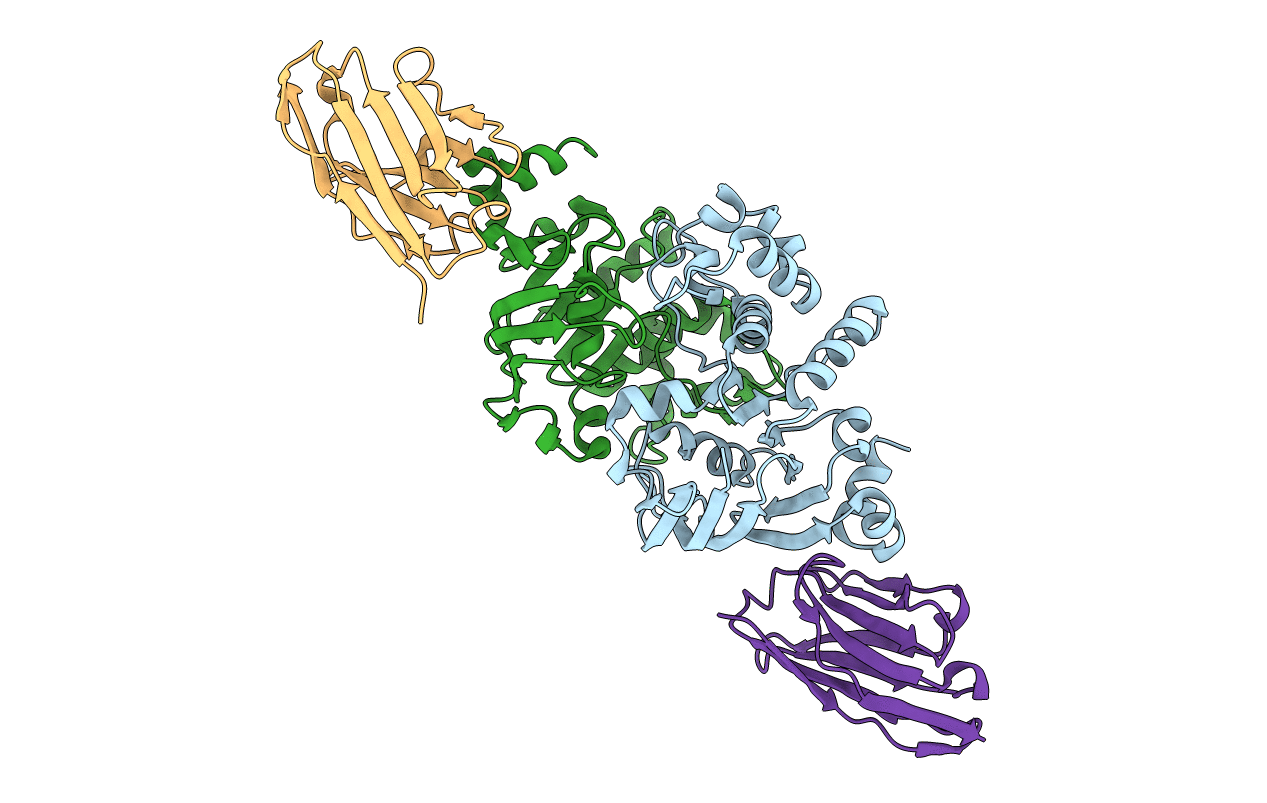 |
Crystal Structure Of Nanobody Nb_Msba#1 In Complex With The Nucleotide Binding Domain Of Msba
Organism: Vicugna pacos, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-03-02 Classification: PROTEIN BINDING |
 |
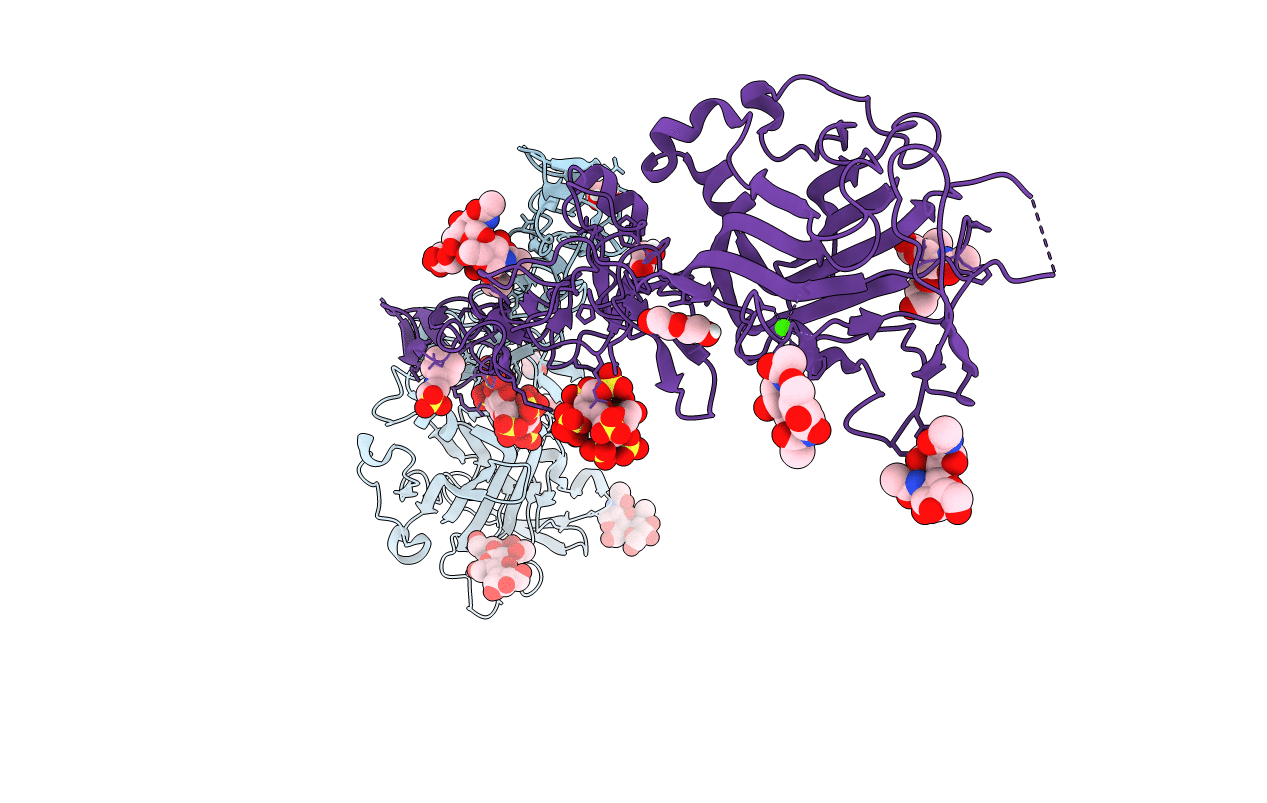
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:5.99 Å Release Date: 2022-02-23 Classification: SIGNALING PROTEIN Ligands: NAG |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:3.21 Å Release Date: 2022-02-23 Classification: SIGNALING PROTEIN Ligands: EDO, PEG, CA, NA, NHE, CL |
 |
Sars-Cov-2 Spike Protein In Complex With Sybody#15 And Sybody#68 In A 3Up Conformation
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-08-04 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Sars-Cov-2 Spike Protein In Complex With Sybody#15 And Sybody#68 In A 1Up/1Up-Out/1Down Conformation
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-08-04 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Sars-Cov-2 Spike Protein In Complex With Sybodyb#15 In A 1Up/1Up-Out/1Down Conformation.
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-08-04 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Sars-Cov-2 Spike Protein In Complex With Sybody#68 In A 2Up/1Flexible Conformation
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-08-04 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Sars-Cov-2 Spike Protein In Complex With Sybody No68 In A 1Up/2Down Conformation
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2021-08-04 Classification: VIRAL PROTEIN |
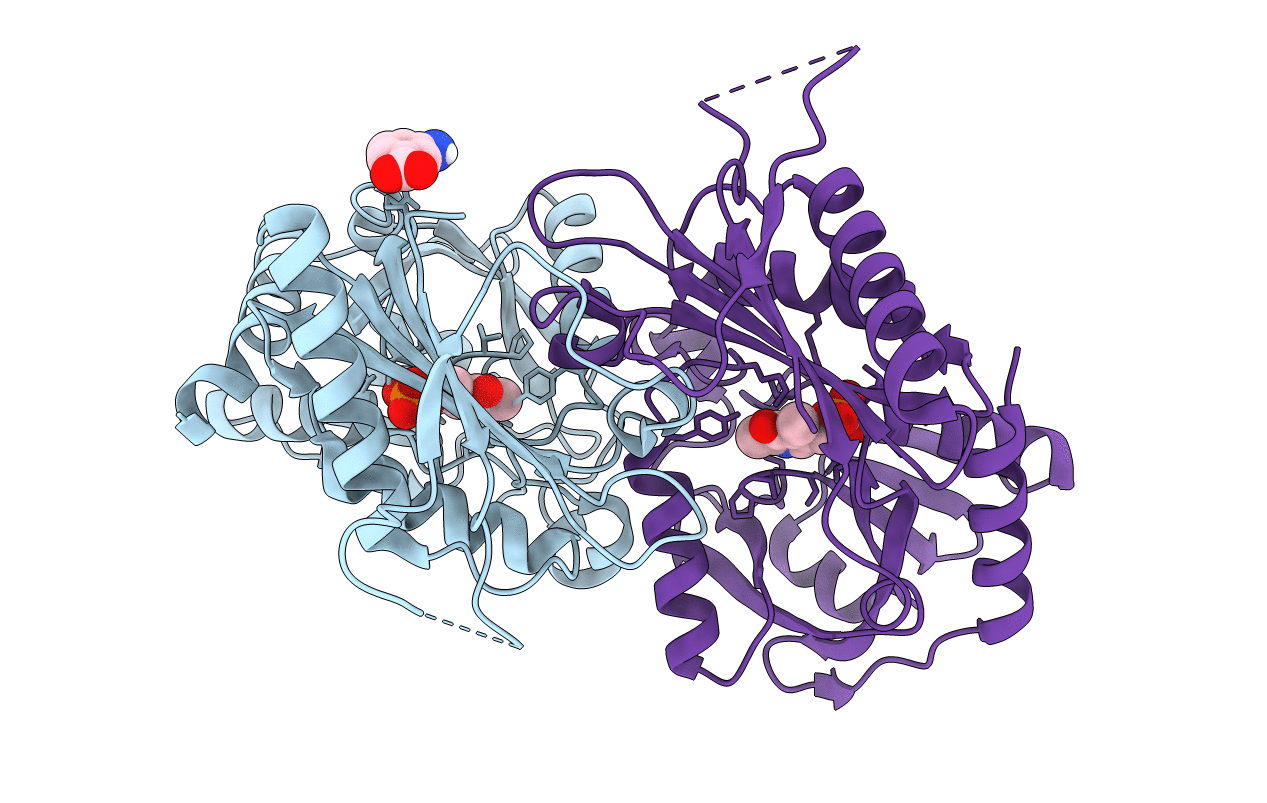 |
Structural Characterisation Of Fold Iv-Transaminase, Cputa1, From Curtobacterium Pusillum
Organism: Curtobacterium pusillum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-12-14 Classification: TRANSFERASE Ligands: PLP, GAB |
 |
Organism: Elizabethkingia meningoseptica
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2015-07-01 Classification: LYASE Ligands: FAD, NA, P6G, PO4 |

