Search Count: 43
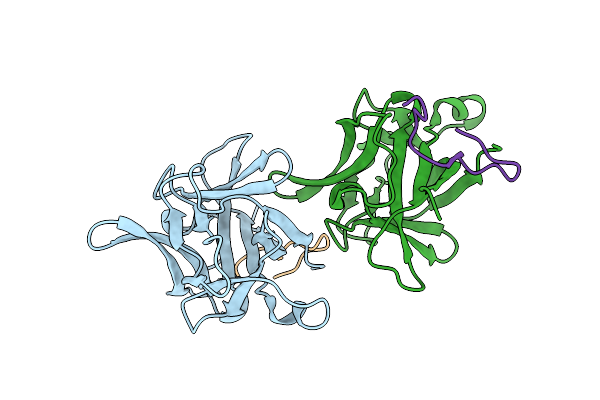 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-05-22 Classification: CYTOKINE |
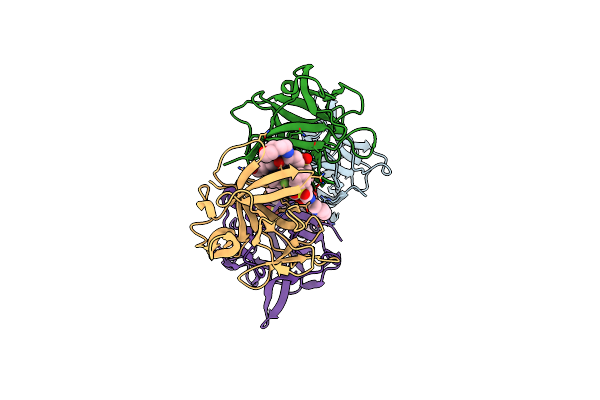 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2024-05-22 Classification: CYTOKINE Ligands: A1H4A |
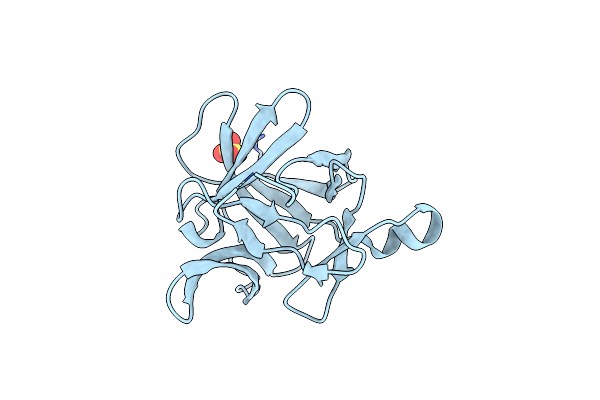 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2024-03-06 Classification: CYTOKINE Ligands: SO4 |
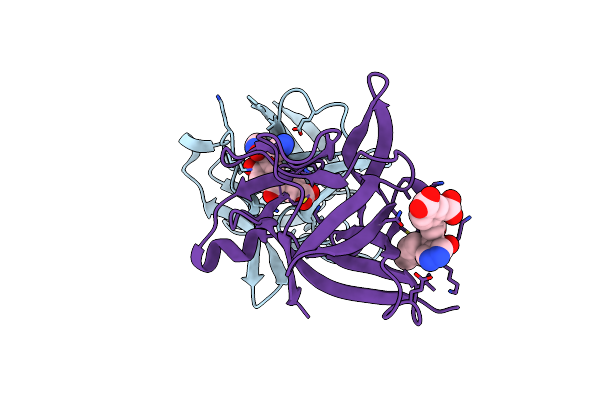 |
Crystal Structure Of Human Il-1Beta In Complex With A Low Molecular Weight Antagonist
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-09-13 Classification: CYTOKINE Ligands: T9C |
 |
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2023-05-10 Classification: Hydrolase, Isomerase Ligands: 12P, ZN, CL |
 |
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2023-05-10 Classification: Hydrolase, Isomerase Ligands: ZN |
 |
Crystal Structure Of T. Maritima Reverse Gyrase With A Minimal Latch, Hexagonal Form
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2023-04-26 Classification: HYDROLASE Ligands: P6G, ZN, SO4, CL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-12-16 Classification: ANTIVIRAL PROTEIN |
 |
Complement Factor B In Complex With 5-Bromo-3-Chloro-N-(4,5-Dihydro-1H-Imidazol-2-Yl)-7-Methyl-1H-Indol-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2020-03-04 Classification: HYDROLASE Ligands: MVZ, SO4 |
 |
Complement Factor B In Complex With (S)-5,7-Dimethyl-4-((2-Phenylpiperidin-1-Yl)Methyl)-1H-Indole
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2020-03-04 Classification: HYDROLASE Ligands: SO4, MVK, ZN |
 |
Complement Factor B In Complex With (-)-4-(1-((5,7-Dimethyl-1H-Indol-4-Yl)Methyl)Piperidin-2-Yl)Benzoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-03-04 Classification: HYDROLASE Ligands: SO4, MVW, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: ZN, 7K1, MPD |
 |
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2016-12-07 Classification: RNA BINDING PROTEIN, HYDROLASE Ligands: SO4 |
 |
Two Different Open Conformations Of The Helicase Core Of The Rna Helicase Hera
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-07-31 Classification: HYDROLASE Ligands: AMP, NA, SO4 |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2013-07-31 Classification: HYDROLASE Ligands: SO4 |
 |
Crystal Structure Of The Rrm Domain Of Rna Helicase Hera From T. Thermophilus In Complex With Gggc Rna
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2013-04-24 Classification: HYDROLASE/RNA |
 |
Crystal Structure Of The R444A / R449A Double Mutant Of The Hera Rna Helicase Rrm Domain
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2013-04-24 Classification: HYDROLASE Ligands: CL, ZN |
 |
Crystal Structure Of The K463A Mutant Of The Rrm Domain Of Rna Helicase Hera From T. Thermophilus
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2013-04-24 Classification: HYDROLASE Ligands: CL |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2013-02-13 Classification: ISOMERASE Ligands: K, TRS |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2012-12-26 Classification: ISOMERASE Ligands: ZN |

