Search Count: 16
 |
Crystal Structure Of A Novel Agarose-Binding Carbohydrate Binding Module Of An Agarase
Organism: Wenyingzhuangia fucanilytica
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-09-11 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Organism: Flammeovirga sp. oc4
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-09-11 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of The Beta,Kappa-Carrageenase Cgbk16A From Wenyingzhuangia Fucanilytica
Organism: Wenyingzhuangia fucanilytica
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2024-07-17 Classification: HYDROLASE |
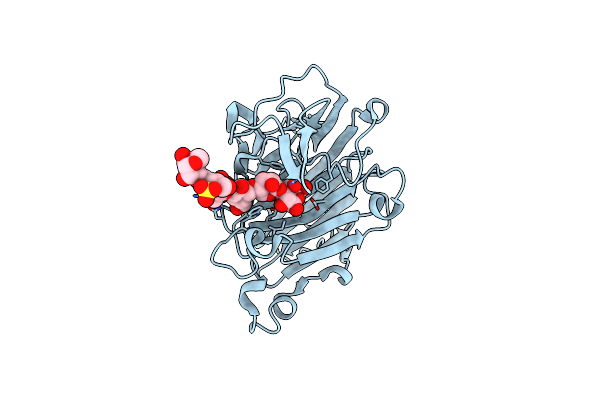 |
Crystal Structure Of The Complex Of The Beta,Kappa-Carrageenase Cgbk16A From Wenyingzhuangia Fucanilytica With An Oligosaccharide Of Furcellaran
Organism: Wenyingzhuangia fucanilytica
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2024-07-17 Classification: HYDROLASE |
 |
Organism: Vibrio breoganii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-06-12 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Aquimarina sp. bl5
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-01-17 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Polaribacter haliotis
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-09-20 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Wenyingzhuangia sp. of219
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2023-02-22 Classification: HYDROLASE Ligands: P6G, CA |
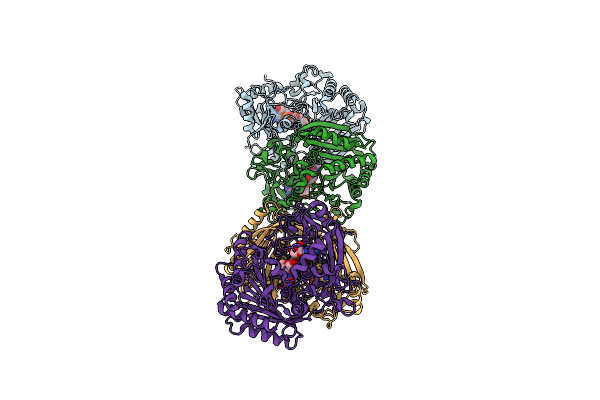 |
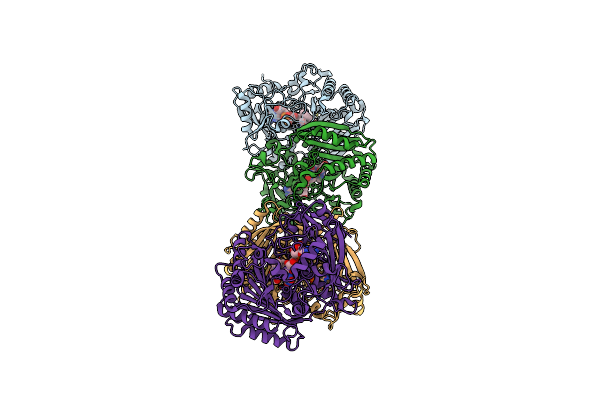
Crystal Structure Of Crmk, A Flavoenzyme Involved In The Shunt Product Recycling Mechanism In Caerulomycin Biosynthesis
Organism: Actinoalloteichus sp. wh1-2216-6
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2017-02-15 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
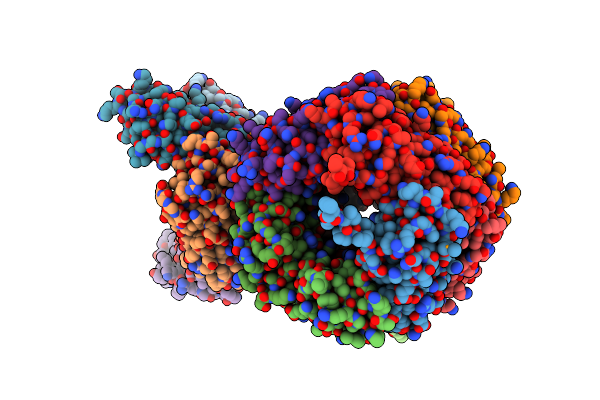
Crystal Structure Of Crmk, A Flavoenzyme Involved In The Shunt Product Recycling Mechanism In Caerulomycin Biosynthesis
Organism: Actinoalloteichus sp. wh1-2216-6
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-02-15 Classification: OXIDOREDUCTASE Ligands: FAD, 67L, 67K |
 |
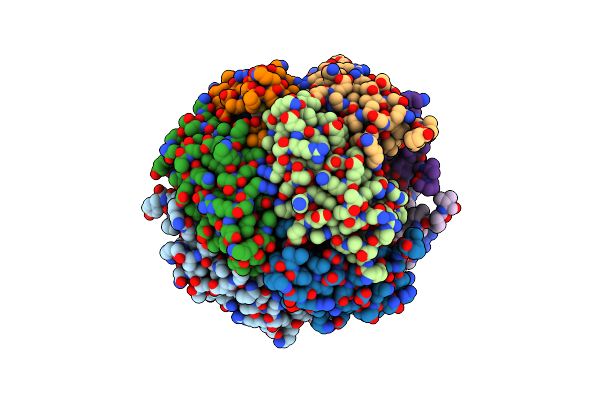
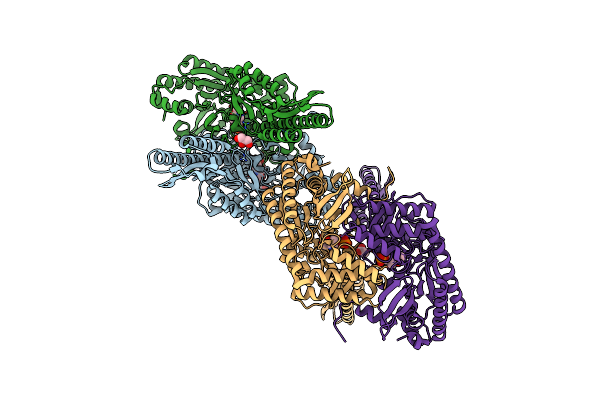
Crystal Structure Of The Covalent Thioimide Intermediate Of The Archaeosine Synthase Quef-Like
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2016-11-09 Classification: TRANSFERASE Ligands: GD1, NA |
 |
Organism: Pyrobaculum calidifontis (strain jcm 11548 / va1)
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2016-11-09 Classification: TRANSFERASE Ligands: SCN, PGE, PG4 |
 |
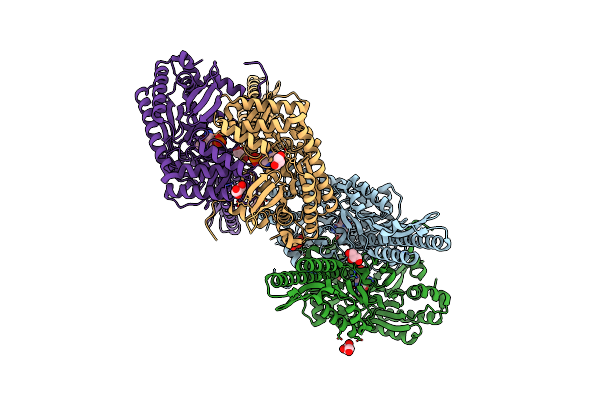
Crystal Structure Of Aminotransferase Crmg From Actinoalloteichus Sp. Wh1-2216-6 In Complex With Plp
Organism: Actinoalloteichus sp. wh1-2216-6
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-08-10 Classification: TRANSFERASE Ligands: PLP, GOL, ACY |
 |
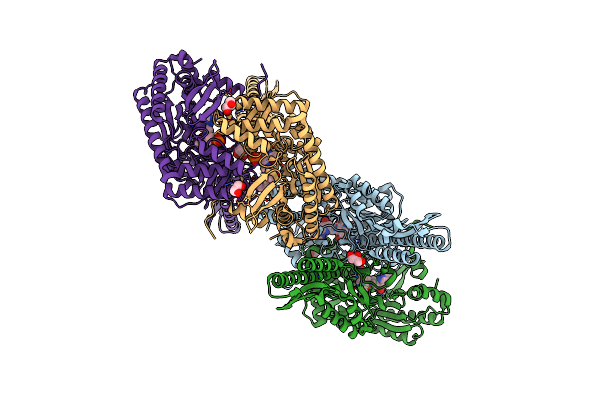
Crystal Structure Of Aminotransferase Crmg From Actinoalloteichus Sp. Wh1-2216-6 In Complex With Pmp
Organism: Actinoalloteichus sp. wh1-2216-6
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2016-08-10 Classification: TRANSFERASE Ligands: PMP, GOL, PGE |
 |
Crystal Structure Of Aminotransferase Crmg From Actinoalloteichus Sp. Wh1-2216-6 In Complex With The Pmp External Aldimine Adduct With Caerulomycin M
Organism: Actinoalloteichus sp. wh1-2216-6
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-08-10 Classification: TRANSFERASE Ligands: 5B6, GOL |
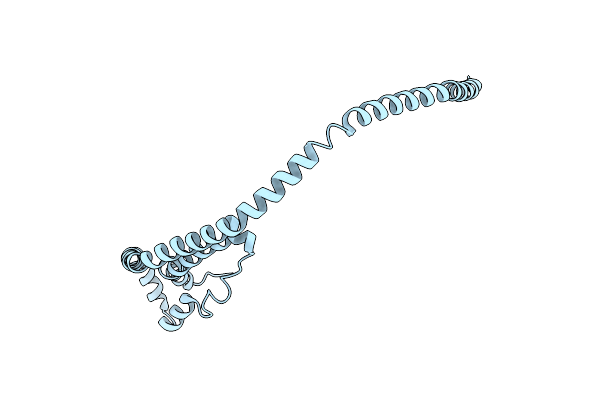 |
2.2 Angstrom Crystal Structure Of C Terminal Truncated Human Apolipoprotein A-I Reveals The Assembly Of Hdl By Dimerization.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2011-09-21 Classification: LIPID TRANSPORT |

