Search Count: 55
 |
Organism: Oplophorus gracilirostris
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-11-16 Classification: OXIDOREDUCTASE Ligands: ACT, FLC |
 |
Organism: Oplophorus gracilirostris
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-11-16 Classification: OXIDOREDUCTASE Ligands: ACT |
 |
2.20A Resolution Structure Of Nanoluc Luciferase With Bound Substrate Analog 3-Methoxy-Furimazine
Organism: Oplophorus gracilirostris
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-11-16 Classification: OXIDOREDUCTASE Ligands: 9Z5 |
 |
1.80A Resolution Structure Of Nanoluc Luciferase With Bound Inhibitor Pc 16026576
Organism: Oplophorus gracilirostris
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-11-16 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: MG, SO4, 9YR, 9Y4, GOL, CL |
 |
1.70A Resolution Structure Of Nanobit Complementation Reporter Complex Of Lgbit And Smbit Subunits
Organism: Oplophorus gracilirostris
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-11-16 Classification: OXIDOREDUCTASE Ligands: GOL |
 |
2.10A Resolution Structure Of Nanobit Complementation Reporter Large Subunit Lgbit
Organism: Oplophorus gracilirostris
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-11-16 Classification: OXIDOREDUCTASE |
 |
Organism: Human respiratory syncytial virus a (strain a2), Human respiratory syncytial virus a, Escherichia phage t2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.74 Å Release Date: 2020-11-11 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-11-11 Classification: IMMUNE SYSTEM |
 |
Low Resolution Crystal Structure (5.5 A) Of The Anthrax Toxin Protective Antigen Heptamer Prepore D425A Mutant
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:5.50 Å Release Date: 2020-10-07 Classification: TOXIN |
 |
2.15A Resolution Structure Of The Cs-B5R Domains Of Human Ncb5Or (Nad+ Form)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-07-17 Classification: OXIDOREDUCTASE Ligands: FAD, NAD |
 |
2.05A Resolution Structure Of The Cs-B5R Domains Of Human Ncb5Or (Nadp+ Form)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-07-17 Classification: OXIDOREDUCTASE Ligands: FAD, NAP |
 |
1.95 A Resolution Structure Of Norovirus 3Cl Protease In Complex With A Triazole-Based Macrocyclic Inhibitor
Organism: Norwalk virus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-11-07 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: DW7 |
 |
2.25 A Resolution Structure Of Norovirus 3Cl Protease In Complex With A Triazole-Based Macrocyclic Inhibitor
Organism: Norwalk virus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-11-07 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 5LH |
 |
1.15 A Resolution Structure Of Norovirus 3Cl Protease In Complex With A Triazole-Based Macrocyclic Inhibitor
Organism: Norwalk virus
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2018-11-07 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: DW4 |
 |
1.50A Resolution Structure Of Catechol O-Methyltransferase (Comt) From Nannospalax Galili
Organism: Nannospalax galili
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-09-12 Classification: TRANSFERASE Ligands: GOL, NA, TRS, PO4 |
 |
1.90A Resolution Structure Of Catechol O-Methyltransferase (Comt) L136M (Hexagonal Form) From Nannospalax Galili
Organism: Nannospalax galili
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-09-12 Classification: TRANSFERASE |
 |
1.70A Resolution Structure Of Catechol O-Methyltransferase (Comt) L136M (Rhombohedral Form) From Nannospalax Galili
Organism: Nannospalax galili
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-09-12 Classification: TRANSFERASE Ligands: SO4 |
 |
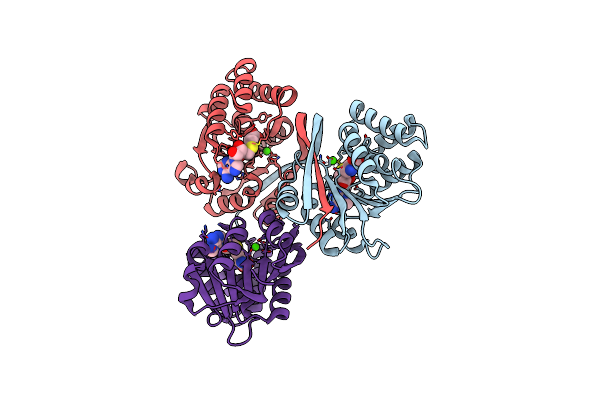
2.15A Resolution Structure Of Sah Bound Catechol O-Methyltransferase (Comt) From Nannospalax Galili
Organism: Nannospalax galili
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2018-09-12 Classification: TRANSFERASE Ligands: CA, SAH |
 |
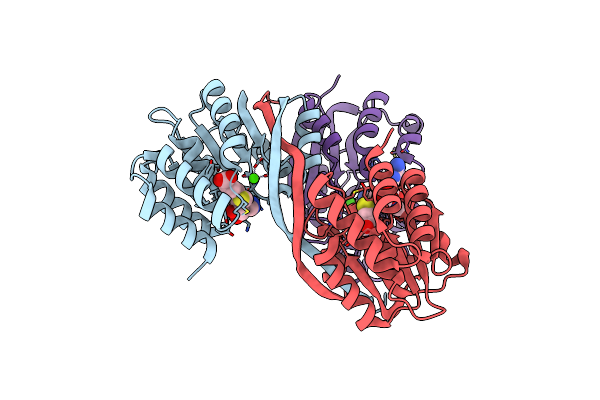
2.25A Resolution Domain Swapped Dimer Structure Of Sah Bound Catechol O-Methyltransferase (Comt) From Nannospalax Galili
Organism: Nannospalax galili
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-09-12 Classification: TRANSFERASE Ligands: CA, SAH |
 |
2.55A Resolution Structure Of Sah Bound Catechol O-Methyltransferase (Comt) L136M From Nannospalax Galili
Organism: Nannospalax galili
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2018-09-12 Classification: TRANSFERASE Ligands: CA, SAH |

