Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
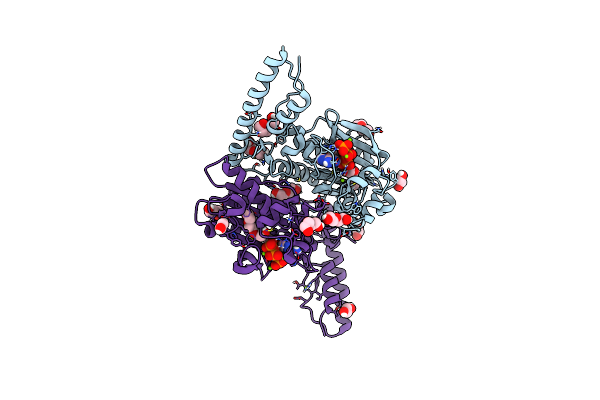 |
Crystal Structure Of Pyrrolysyl-Trna Synthetase From Methanomethylophilus Alvus Engineered For 3-Methyl-L-Histidine, Bound To Amppnp
Organism: Candidatus methanomethylophilus alvus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2023-07-19 Classification: LIGASE Ligands: ANP, PEG, PGE, EDO, MG |
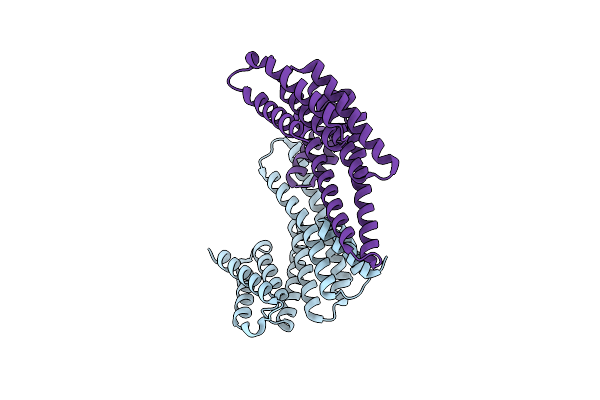 |
The Crystal Structure Of 14-3-3 Beta Containing 3-Nitrotyrosine At Position Y130
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-01-25 Classification: PROTEIN BINDING |
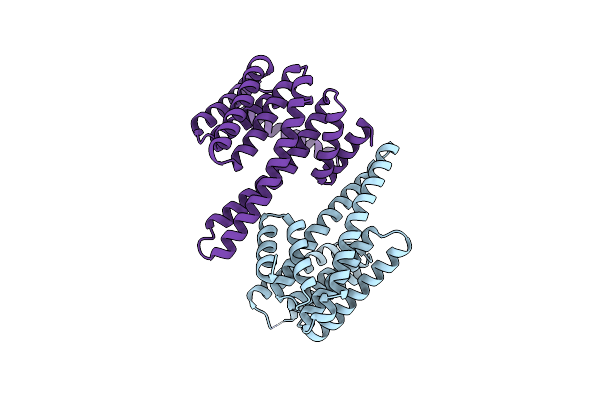 |
The Crystal Structure Of 14-3-3 Beta Containing 3-Nitrotyrosine At Position Y213
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-01-25 Classification: PROTEIN BINDING |
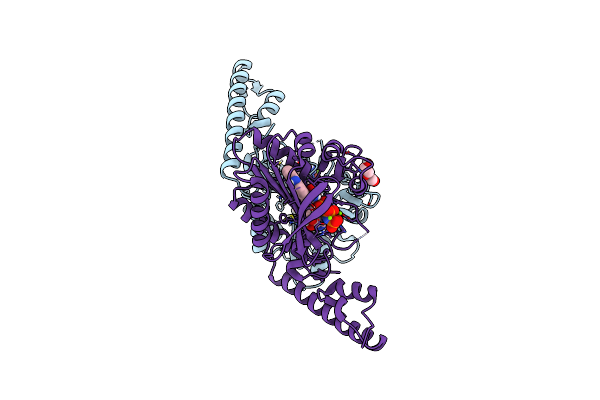 |
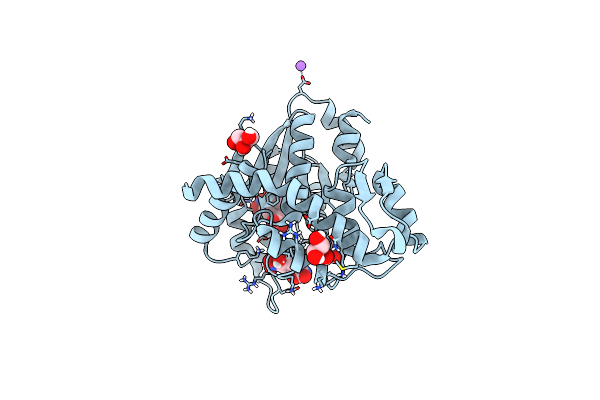
Crystal Structure Of Pyrrolysyl-Trna Synthetase From Methanomethylophilus Alvus Engineered For Acridone Amino Acid (Rs1) Bound To Amppnp And Acridone
Organism: Candidatus methanomethylophilus alvus
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2022-12-07 Classification: LIGASE Ligands: ANP, T7Q, MG, GOL, PEG |
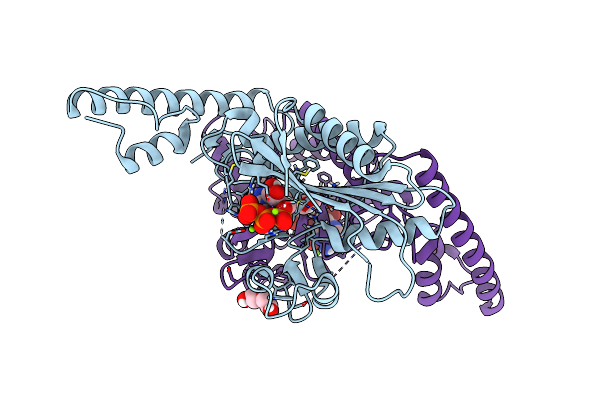 |
Crystal Structure Of Pyrrolysyl-Trna Synthetase From Methanomethylophilus Alvus Engineered For Acridone Amino Acid (Rs1) Bound To Atp And Acridone After 24 Hours Of Crystal Growth
Organism: Candidatus methanomethylophilus alvus
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2022-12-07 Classification: LIGASE Ligands: ATP, T7Q, PEG, MG, GOL |
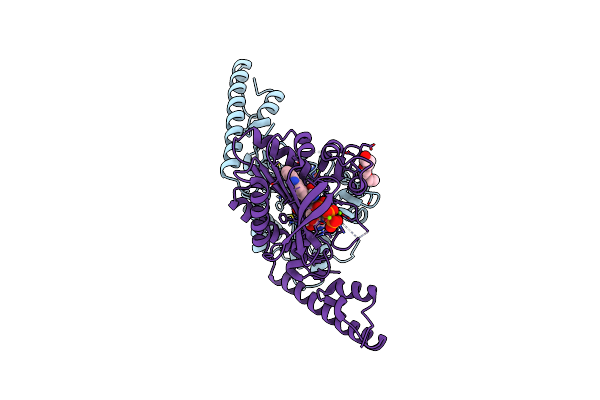 |
Crystal Structure Of Pyrrolysyl-Trna Synthetase From Methanomethylophilus Alvus Engineered For Acridone Amino Acid (Rs1) Bound To Atp And Acridone After 2- Weeks Of Crystal Growth
Organism: Candidatus methanomethylophilus alvus
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2022-12-07 Classification: LIGASE Ligands: ATP, T7Q, PG4, MG, GOL |
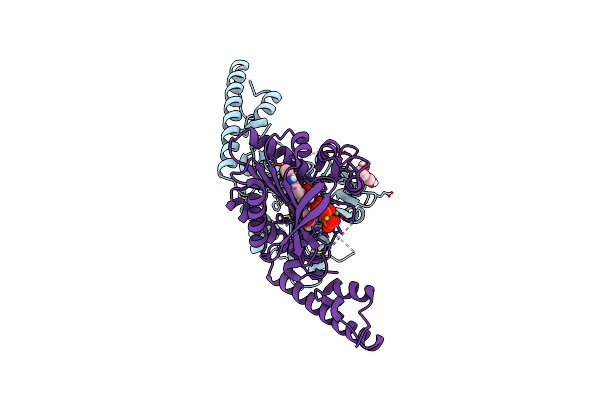 |
Crystal Structure Of Pyrrolysyl-Trna Synthetase From Methanomethylophilus Alvus Engineered For Acridone Amino Acid (Ast) Bound To Atp And Acridone
Organism: Candidatus methanomethylophilus alvus
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2022-12-07 Classification: LIGASE Ligands: ATP, T7Q, MG, GOL, AMP, PEG |
 |
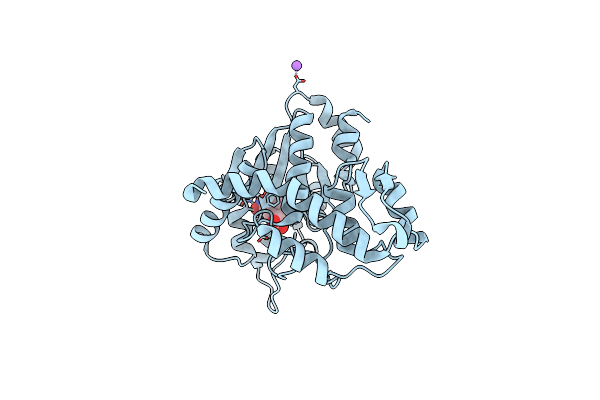
Crystal Structure Of 3Rd-Generation Mj 3-Nitro-Tyrosine Trna Synthetase ("A7") Bound To 3-Nitro-Tyrosine
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-07-01 Classification: LIGASE Ligands: GOL, NA, NIY |
 |
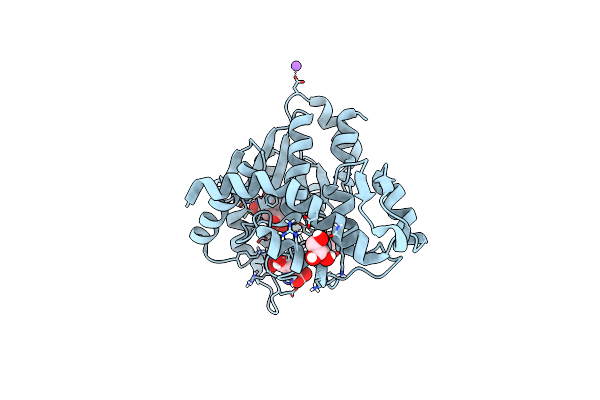
Crystal Structure Of Mj 3-Nitro-Tyrosine Trna Synthetase (5B) C70A Variant Bound To 3-Nitro-Tyrosine
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-07-01 Classification: LIGASE |
 |
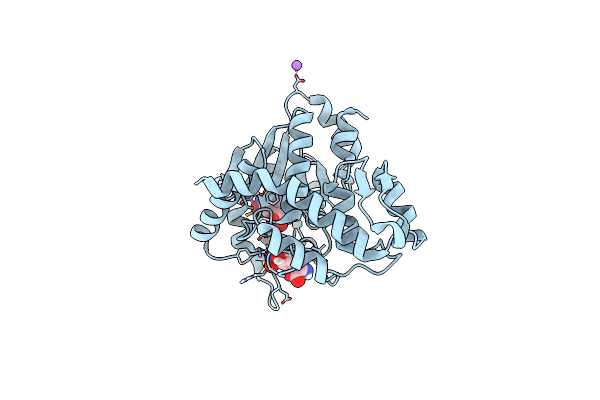
Crystal Structure Of Mj 3-Nitro-Tyrosine Trna Synthetase (5B) S158C Variant Bound To 3-Nitro-Tyrosine
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-07-01 Classification: LIGASE Ligands: GOL, NA, NIY |
 |
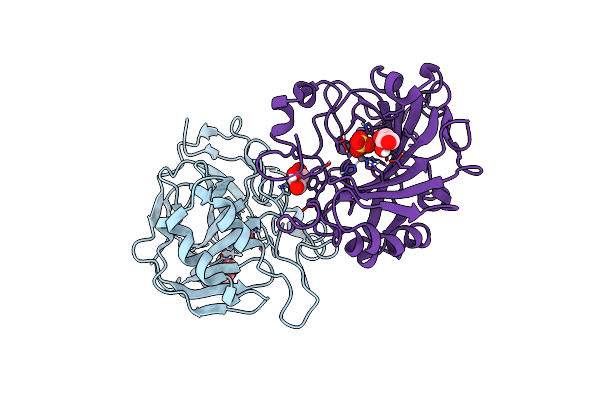
Crystal Structure Of Mj 3-Nitro-Tyrosine Trna Synthetase (5B) C70A/S158C Variant Bound To 3-Nitro-Tyrosine
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-07-01 Classification: LIGASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-10-02 Classification: LYASE Ligands: ZN, SO4, GOL |
 |
Thermostable Variant Of Human Carbonic Anhydrase With Ordered Tetrazine 2.0 At Site 233
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.01 Å Release Date: 2019-10-02 Classification: LYASE Ligands: ACT, ZN |
 |
Thermostable Variant Of Human Carbonic Anhydrase With Disordered Tetrazine 2.0 Reacted With Strained Trans-Cyclooctene At Site 233
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2019-10-02 Classification: LYASE Ligands: ZN |
 |
Thermostable Variant Of Human Carbonic Anhydrase Ii With Disordered Tetrazine 2.0 At Site 233
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2019-10-02 Classification: LYASE Ligands: ZN |
 |
Thermostable Variant Of Human Carbonic Anhydrase With Tetrazine 2.0 At Site 186 Reacted With Stco In Crystallo
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-10-02 Classification: LYASE Ligands: MOH, SO4, GOL, ZN |
 |
Organism: Streptomyces fradiae
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2019-03-13 Classification: TRANSFERASE Ligands: SAH, TLO |
 |
Organism: Streptomyces fradiae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-03-13 Classification: TRANSFERASE Ligands: SAH, TLO, EDO |
 |
Organism: Streptomyces fradiae
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2019-03-13 Classification: TRANSFERASE Ligands: SAH, TLO, CL |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2018-06-20 Classification: HYDROLASE Ligands: HED |