Search Count: 38
 |
The Structure Of Archaeal Inorganic Pyrophosphatase In Complex With Pyrophosphate
Organism: Thermococcus thioreducens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-02-08 Classification: HYDROLASE Ligands: CA, EDO, MRD, POP, NA |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-02-26 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:4.50 Å Release Date: 2014-02-26 Classification: IMMUNE SYSTEM Ligands: MAN, NAG |
 |
The Structure Of Manganese Bound Thermococcus Thioreducens Inorganic Pyrophosphatase
Organism: Thermococcus thioreducens
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2012-03-28 Classification: HYDROLASE Ligands: MN |
 |
The Structure Of Calcium Bound Thermococcus Thioreducens Inorganic Pyrophosphatase At 298K
Organism: Thermococcus thioreducens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2012-03-28 Classification: HYDROLASE Ligands: CA, MRD, MPD |
 |
The Structure Of Thermococcus Thioreducens' Inorganic Pyrophosphatase Bound To Sulfate
Organism: Thermococcus thioreducens
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2012-03-28 Classification: HYDROLASE Ligands: SO4, NH4 |
 |
The Structure Of Inorganic Pyrophosphatase From Thermococcus Thioreducens In Complex With Magnesium And Sulfate
Organism: Thermococcus thioreducens
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2012-01-04 Classification: HYDROLASE Ligands: MG, SO4 |
 |
Organism: Homo sapiens, Helianthus annuus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-08-10 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GSH |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2011-08-03 Classification: HYDROLASE Ligands: SO4, EDO, BEN, GSH |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2010-04-21 Classification: HYDROLASE Ligands: SO4, ABV |
 |
Crystal Structure Of Complex Of Urokinase And A Upain-1 Variant(W3A) In Ph4.6 Condition
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2010-04-07 Classification: Hydrolase/Hydrolase inhibitor |
 |
Crystal Structure Of A Triacylglycerol Lipase From Penicillium Expansum At 1.3
Organism: Penicillium expansum
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2010-02-23 Classification: HYDROLASE Ligands: SO4, PEG, 1PE |
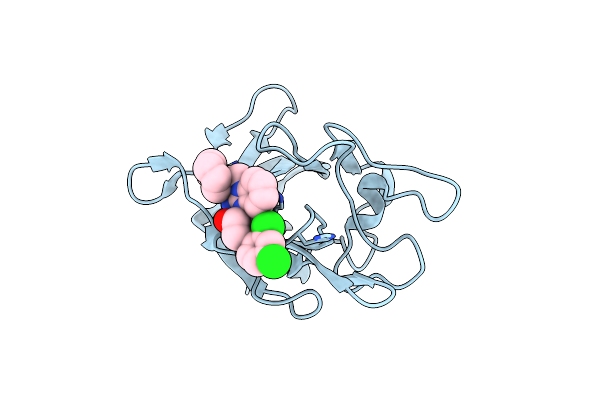 |
Factor Viii Trp2313-His2315 Segment Is Involved In Membrane Binding As Shown By Crystal Structure Of Complex Between Factor Viii C2 Domain And An Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2010-01-19 Classification: BLOOD CLOTTING Ligands: 768 |
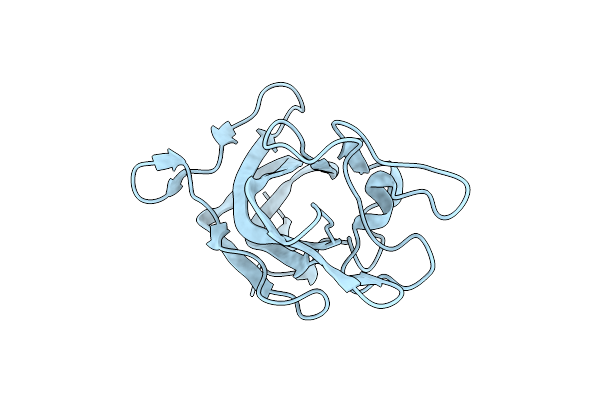 |
Factor Viii Trp2313-His2315 Segment Is Involved In Membrane Binding As Shown By Crystal Structure Of Complex Between Factor Viii C2 Domain And An Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2010-01-19 Classification: BLOOD CLOTTING |
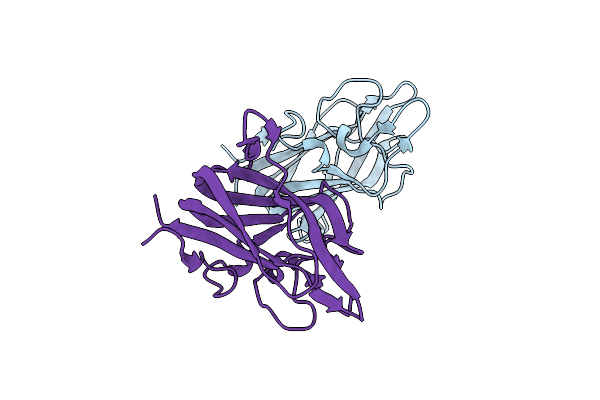 |
Factor Viii Trp2313-His2315 Segment Is Involved In Membrane Binding As Shown By Crystal Structure Of Complex Between Factor Viii C2 Domain And An Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2010-01-19 Classification: BLOOD CLOTTING |
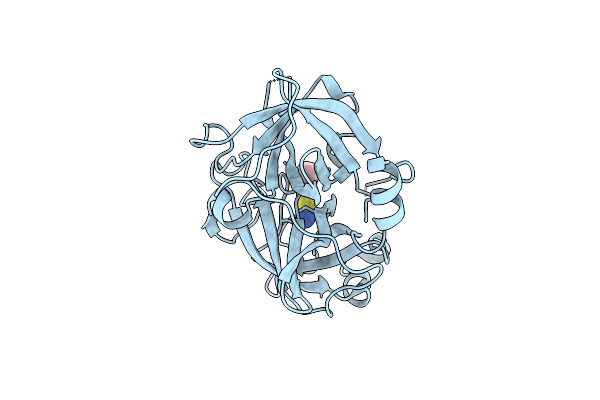 |
The Crystal Structures Of 2-Aminobenzothiazole-Based Inhibitors In Complexes With Urokinase-Type Plasminogen Activator
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2009-12-01 Classification: HYDROLASE Ligands: 2BS |
 |
X-Ray Crystallographic Structure Of Inorganic Pyrophosphatase At 298K From Archaeon Thermococcus Thioreducens
Organism: Thermococcus thioreducens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2009-08-18 Classification: HYDROLASE Ligands: MRD, CA, MPD, ACE |
 |
Crystal Structure Of Human Serum Albumin Complexed With Myristic Acid And Lysophosphatidylethanolamine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2009-04-28 Classification: STRUCTURAL PROTEIN Ligands: MYR, LPX |
 |
Crystal Structure Of Ubiquitin-Conjugating Enzyme E2-25Kda (Huntington Interacting Protein 2) M172A Mutant Crystallized At Ph 8.5
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2008-11-25 Classification: LIGASE Ligands: PG4, BME, TRS, CA |
 |
Atomic Resolution Structure Of Cucurmosin, A Novel Type 1 Rip From The Sarcocarp Of Cucurbita Moschata
Organism: Cucurbita moschata
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2008-10-07 Classification: TRANSLATION Ligands: PO4, EDO |

