Search Count: 79
 |
Organism: Acidothermus cellulolyticus
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: DNA BINDING PROTEIN/DNA/RNA |
 |
Crystal Structure Of Udp-N-Acetylenolpyruvoylglucosamine Reductase (Murb) From Brucella Ovis
Organism: Brucella ovis atcc 25840
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2025-01-22 Classification: OXIDOREDUCTASE Ligands: SO4, FAD, EPU |
 |
Room Temperature Structure Of Fad-Containing Ferrodoxin-Nadp Reductase From Brucella Ovis At Euxfel
Organism: Brucella ovis atcc 25840
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Room Temperature Structure Of Fad-Containing Ferrodoxin-Nadp Reductase From Brucella Ovis At Lcls
Organism: Brucella ovis atcc 25840
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-05-29 Classification: FLAVOPROTEIN Ligands: FAD, NAD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-05-29 Classification: FLAVOPROTEIN Ligands: FAD, EPE |
 |
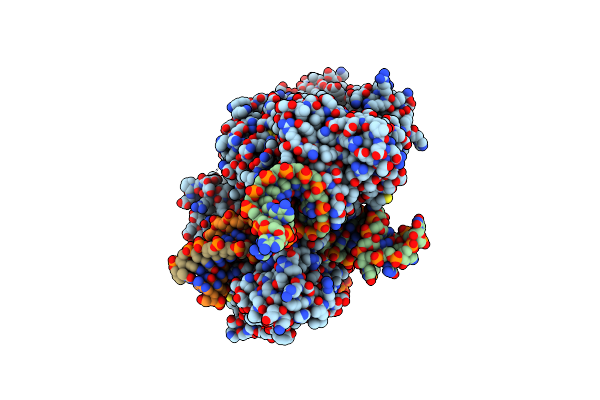
Structure Of Acidothermus Cellulolyticus Cas9 Ternary Complex (Cleavage Intermediate 2)
Organism: Acidothermus cellulolyticus 11b, Acidothermus cellulolyticus
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: MG |
 |
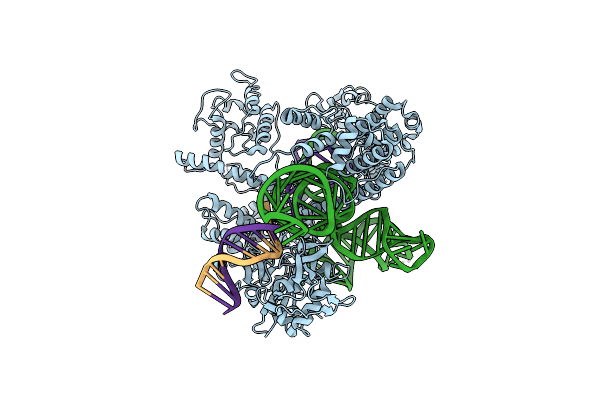
Structure Of Acidothermus Cellulolyticus Cas9 Ternary Complex (Cleavage Intermediate 1)
Organism: Acidothermus cellulolyticus 11b, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: MG |
 |
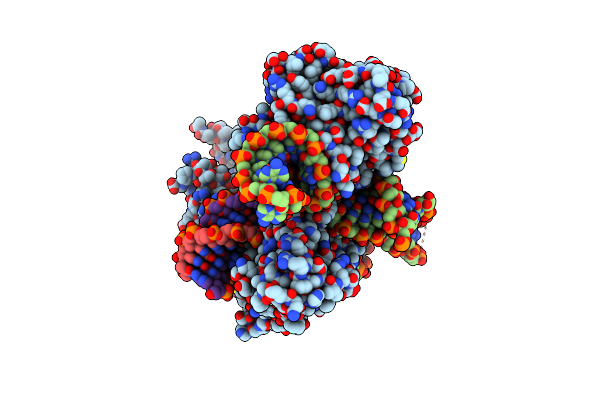
Structure Of Acidothermus Cellulolyticus Cas9 Ternary Complex (Pre-Cleavage)
Organism: Acidothermus cellulolyticus 11b, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: MG |
 |
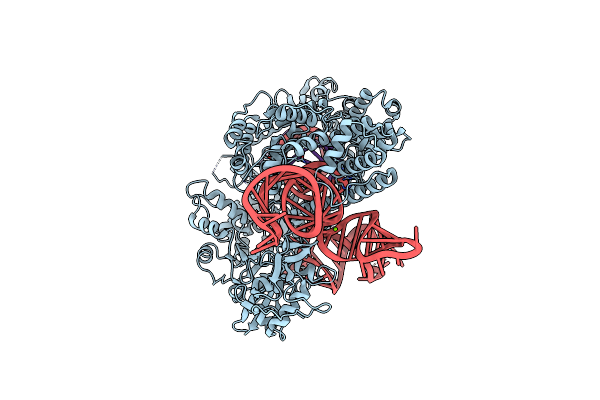
Structure Of Acidothermus Cellulolyticus Cas9 Ternary Complex (Post-Cleavage 2)
Organism: Acidothermus cellulolyticus 11b, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: MG |
 |
Structure Of Acidothermus Cellulolyticus Cas9 Ternary Complex (Target Bound)
Organism: Acidothermus cellulolyticus 11b, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: MG |
 |
Structure Of Acidothermus Cellulolyticus Cas9 Ternary Complex (Post-Cleavage 1)
Organism: Acidothermus cellulolyticus 11b, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: MG |
 |
Crystal Structure A Thioredoxin Reductase From Gloeobacter Violaceus Bound To Its Electron Donor
Organism: Gloeobacter violaceus (strain atcc 29082 / pcc 7421)
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2021-07-28 Classification: FLAVOPROTEIN Ligands: FAD, ACT, PEG, PG4, PGE, FES |
 |
Crystal Structure Of Peruvianin-I (Cysteine Peptidase From Thevetia Peruviana Latex)
Organism: Thevetia peruviana
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-05-06 Classification: HYDROLASE |
 |
Crystal Structure Of Fad-Containing Ferredoxin-Nadp Reductase From Brucella Ovis
Organism: Brucella ovis atcc 25840
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2019-08-21 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
X-Ray Structure Of The Ferredoxin-Nadp Reductase From Brucella Ovis In Complex With Nadp
Organism: Brucella ovis (strain atcc 25840 / 63/290 / nctc 10512)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-08-21 Classification: OXIDOREDUCTASE Ligands: FAD, NAP, SO4 |
 |
Crystal Structure Of Aryl-Alcohol Oxidase From Pleurotus Eryngii In Complex With P-Anisic Acid
Organism: Pleurotus eryngii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-11-01 Classification: OXIDOREDUCTASE Ligands: ANN, FAD, GOL |
 |
Organism: Nostoc sp. (strain atcc 29151 / pcc 7119)
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2017-08-02 Classification: ELECTRON TRANSPORT Ligands: FMN |
 |
Structure Of Thermus Thermophilus L-Proline Dehydrogenase Lacking Alpha Helices A, B And C
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-03-15 Classification: OXIDOREDUCTASE Ligands: FMN |
 |
Organism: Corynebacterium ammoniagenes
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2016-11-30 Classification: TRANSFERASE Ligands: PPV, SO4 |

