Search Count: 51
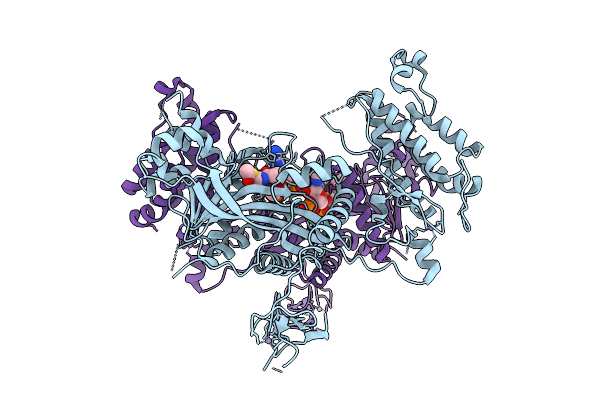 |
Crystal Structure Of Crebbp Histone Acetyltransferase Domain In Complex With Acetyl-Coenzyme A
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: ZN, ACO |
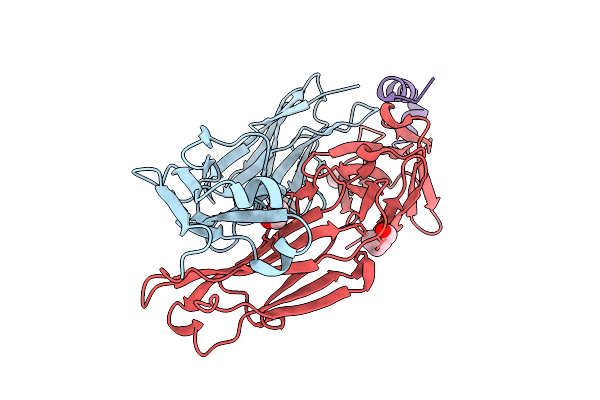 |
Organism: Homo sapiens, Hepatitis b virus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: IMMUNE SYSTEM Ligands: EDO, PEG |
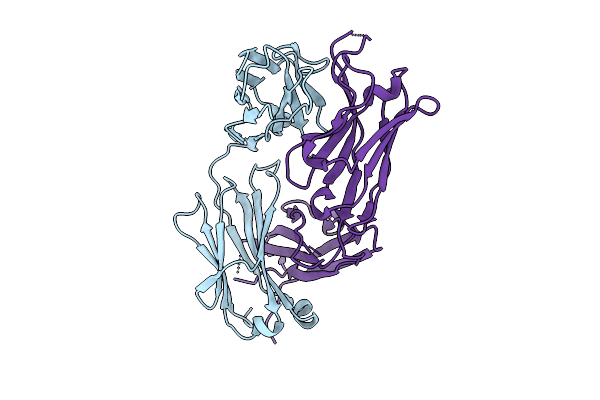 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: IMMUNE SYSTEM |
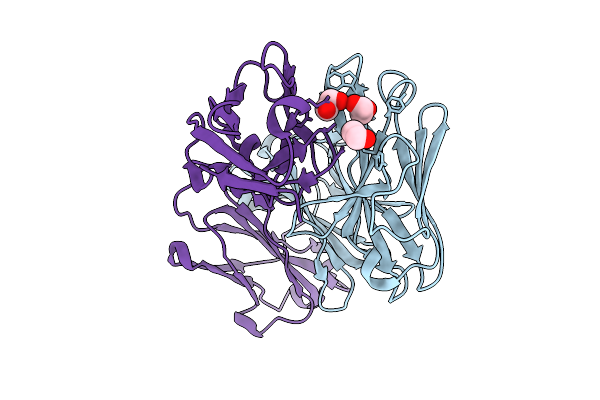 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: IMMUNE SYSTEM Ligands: EDO |
 |
Crystal Structure Of Human Crebbp Histone Acetyltransferase Domain In Complex With Propionyl- Coenzyme A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: 1VU, SO4 |
 |
Crystal Structure Of Human Crebbp Histone Acetyltransferase Domain In Complex With A Bisubstrate Inhibitor, Lys-Coa
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: 01K, EDO |
 |
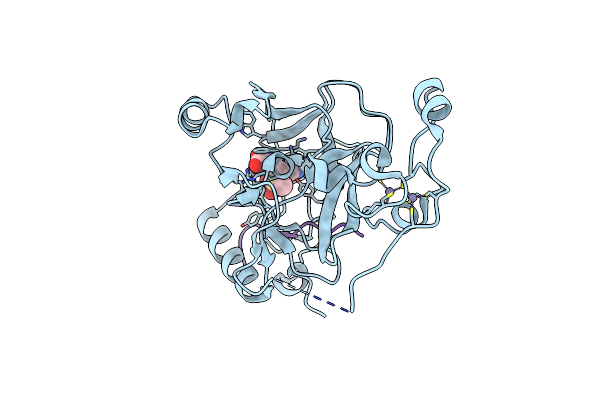
Structure Of Human Setd2 T1663M Mutant In Complex With Sam And H3K36M Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: ZN, SAM |
 |
Structure Of Human Setd2 L1609P Mutant In Complex With Sam And H3K36M Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2025-03-05 Classification: TRANSFERASE Ligands: ZN, SAM |
 |
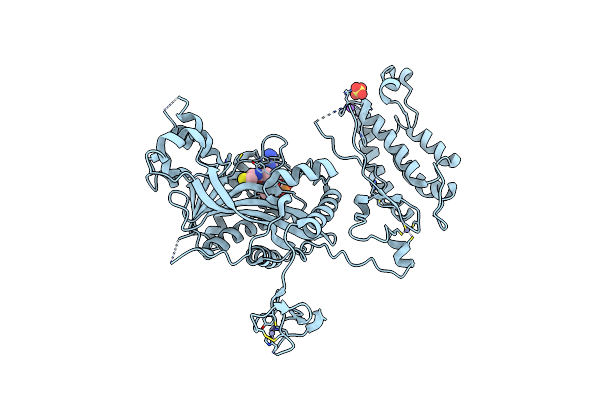
Structure Of The Lysine Methyltransferase Setd2 In Complex With A Peptide Derived From Human Tyrosine Kinase Ack1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-04-24 Classification: TRANSFERASE Ligands: ZN, SAM |
 |
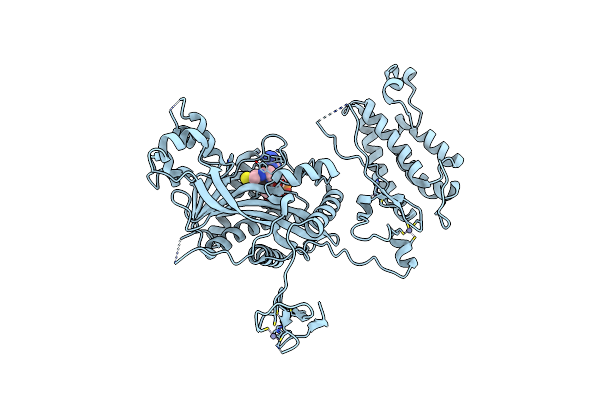
Crystal Structure Of Crebbp Histone Acetyltransferase Domain In Complex With Coenzyme A
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2024-04-03 Classification: TRANSFERASE Ligands: SO4, ZN, COA, NA |
 |
Crystal Structure Of Crebbp-R1446C Histone Acetyltransferase Domain In Complex With Coenzyme A
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2024-03-06 Classification: TRANSFERASE Ligands: ZN, COA |
 |
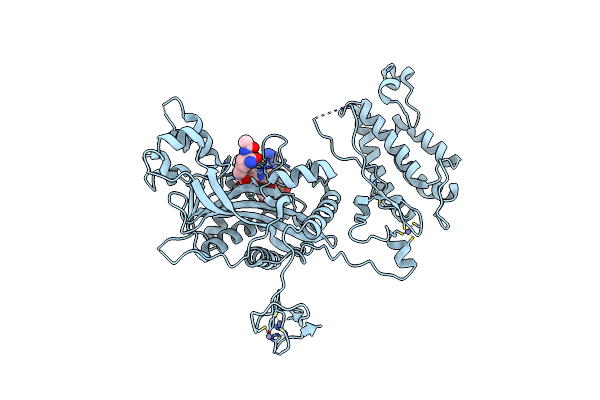
Crystal Structure Of Crebbp-Y1482N Histone Acetyltransferase Domain In Complex With Coenzyme A
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2024-03-06 Classification: TRANSFERASE Ligands: ZN, COA |
 |
Crystal Structure Of Crebbp-R1446C Histone Acetyltransferase Domain In Complex With A Bisubstrate Inhibitor, Lys-Coa
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2024-03-06 Classification: TRANSFERASE Ligands: ZN, 01K |
 |
Crystal Structure Of Crebbp-Y1503C Histone Acetyltransferase Domain In Complex With Coenzyme A
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-03-06 Classification: TRANSFERASE Ligands: ZN, COA |
 |
Crystal Structure Of Crebbp-Y1482N Histone Acetyltransferase Domain In Complex With A Bisubstrate Inhibitor, Lys-Coa
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2024-03-06 Classification: TRANSFERASE Ligands: ZN, 01K |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2021-08-18 Classification: SIGNALING PROTEIN Ligands: PO4 |
 |
Structural Insights Into Cubane-Modified Aptamer Recognition Of A Malaria Biomarker
Organism: Plasmodium vivax, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2020-07-15 Classification: DNA Ligands: O0Q |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2017-06-07 Classification: TRANSFERASE Ligands: SO4, ATP, MG |
 |
Crystal Structure Of The Desk-Desr Complex In The Phosphotransfer State With Low Mg2+ (20 Mm)
Organism: Bacillus subtilis, Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-12-21 Classification: TRANSFERASE Ligands: ACP, MG, K |
 |
Crystal Structure Of The Desk-Desr Complex In The Phosphotransfer State With High Mg2+ (150 Mm)
Organism: Bacillus subtilis, Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2016-12-21 Classification: TRANSFERASE/GENE REGULATION Ligands: ACP, MG, K |

