Search Count: 19
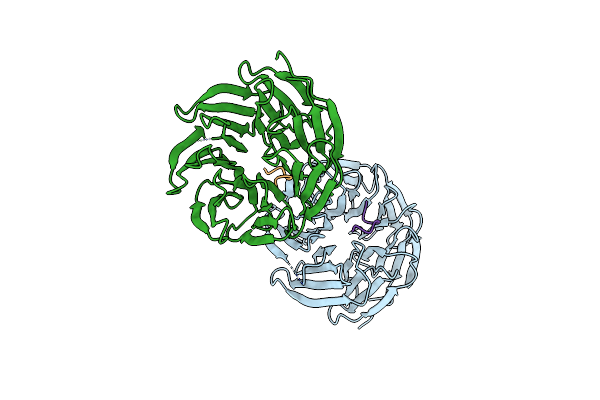 |
X-Ray Structure Of Wdr5Delta24 Bound To The Kaposi'S Sarcoma Herpesvirus Lana Win Motif Peptide
Organism: Homo sapiens, Human herpesvirus 8 type m
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-12-08 Classification: TRANSFERASE |
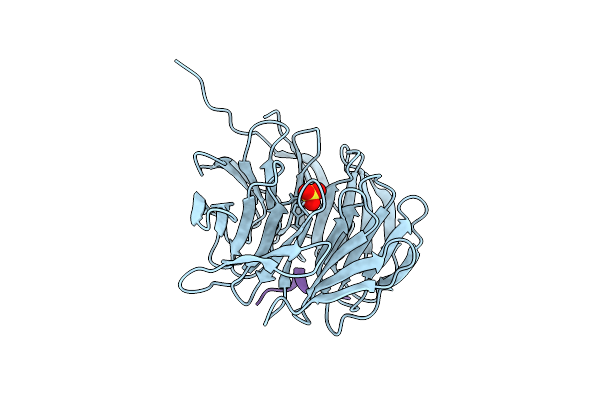 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2021-12-08 Classification: TRANSFERASE Ligands: SO4 |
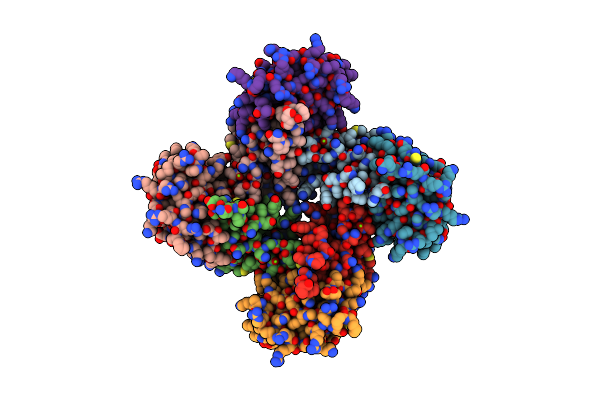 |
Kshv Lana (Orf73) C-Terminal Domain, Open Non-Ring Conformation: Orthorhombic Crystal Form
Organism: Human herpesvirus 8
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2015-10-28 Classification: DNA BINDING PROTEIN Ligands: MG |
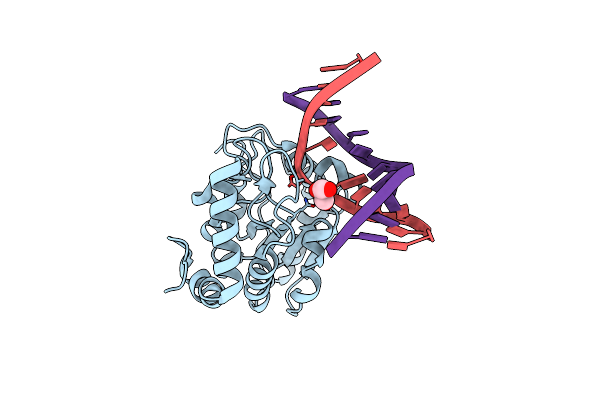 |
Crystal Structure Determination Of Uracil-Dna N-Glycosylase (Ung) From Deinococcus Radiodurans In Complex With Dna - New Insights Into The Role Of The Leucine-Loop For Damage Recognition And Repair
Organism: Deinococcus radiodurans, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2015-08-12 Classification: HYDROLASE/DNA Ligands: CL, GOL |
 |
The Importance Of The Abn2 Calcium Cluster In The Endo-1,5- Arabinanase Activity From Bacillus Subtilis
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-03-05 Classification: HYDROLASE Ligands: NI, TRS |
 |
Crystal Structure Of Mhv-68 Latency-Associated Nuclear Antigen (Lana) C-Terminal Dna Binding Domain
Organism: Murid herpesvirus 4
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-10-30 Classification: VIRAL PROTEIN Ligands: PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2011-10-05 Classification: HYDROLASE Ligands: ATP |
 |
Crystal Structure Of The Full-Length Sorbitol Operon Regulator Sorc From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2009-05-05 Classification: TRANSCRIPTION Ligands: GOL, MPD, CL, MRD |
 |
Organism: Vaccinia virus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2007-05-22 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2006-10-05 Classification: HYDROLASE Ligands: C5P, MG, CA |
 |
Organism: Escherichia coli, Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2006-10-05 Classification: HYDROLASE Ligands: MG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-07-17 Classification: HYDROLASE Ligands: EDO, CA |
 |
Crystal Structures Of Penicillin Acylase Enzyme-Substrate Complexes: Structural Insights Into The Catalytic Mechanism
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2002-01-04 Classification: ANTIBIOTIC RESISTANCE Ligands: EDO, CA |
 |
Crystal Structures Of Penicillin Acylase Enzyme-Substrate Complexes: Structural Insights Into The Catalytic Mechanism
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2002-01-04 Classification: HYDROLASE Ligands: EDO, CA |
 |
Crystal Structures Of Penicillin Acylase Enzyme-Substrate Complexes: Structural Insights Into The Catalytic Mechanism
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2001-11-28 Classification: HYDROLASE Ligands: EDO, CA, PNN |
 |
Crystal Structures Of Penicillin Acylase Enzyme-Substrate Complexes: Structural Insights Into The Catalytic Mechanism
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2001-11-28 Classification: HYDROLASE Ligands: CA, SOX |
 |
Crystal Structures Of Penicillin Acylase Enzyme-Substrate Complexes: Structural Insights Into The Catalytic Mechanism
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-11-28 Classification: HYDROLASE Ligands: EDO, CA, SOX |
 |
Organism: Lysinibacillus sphaericus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2000-07-26 Classification: HYDROLASE Ligands: DTD |
 |
Organism: Lysinibacillus sphaericus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 1999-11-15 Classification: HYDROLASE |

