Search Count: 118
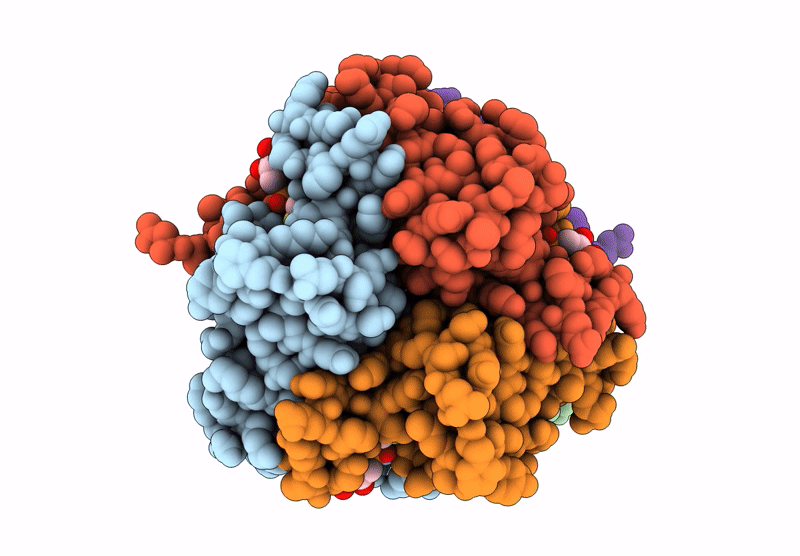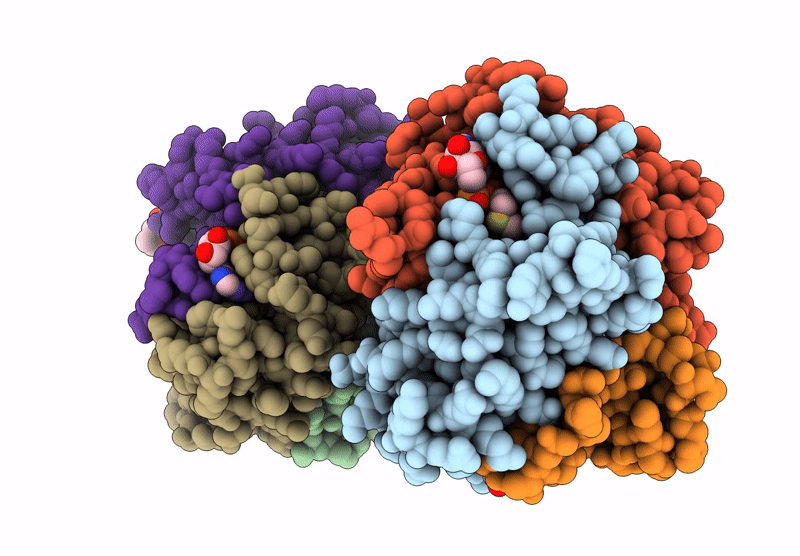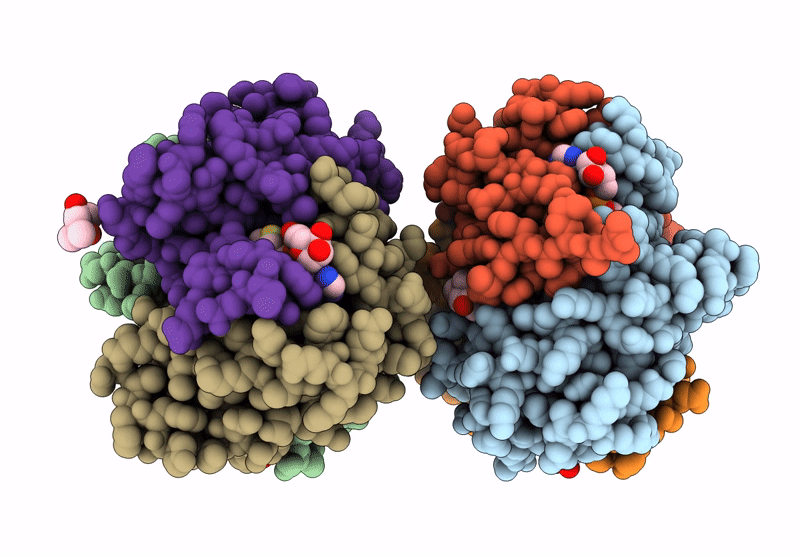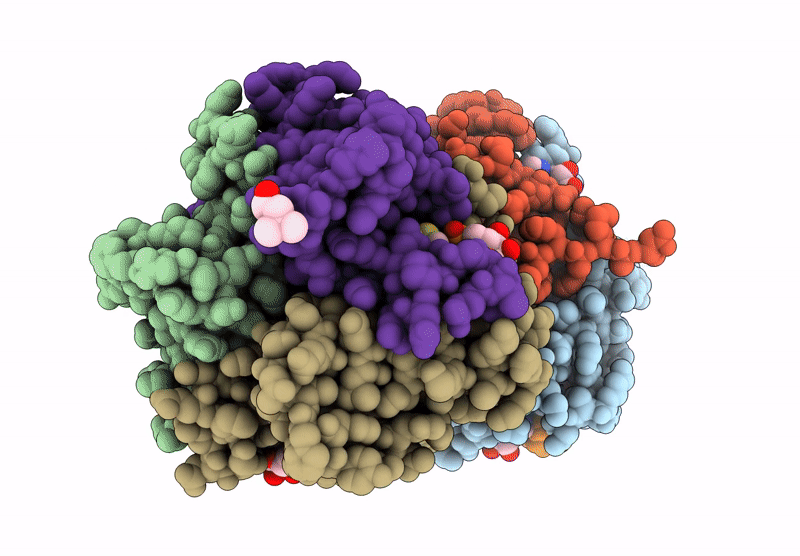 |
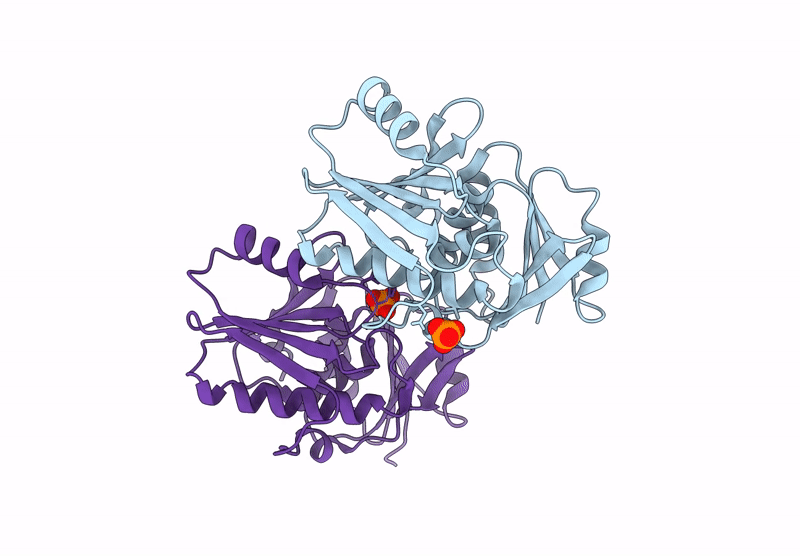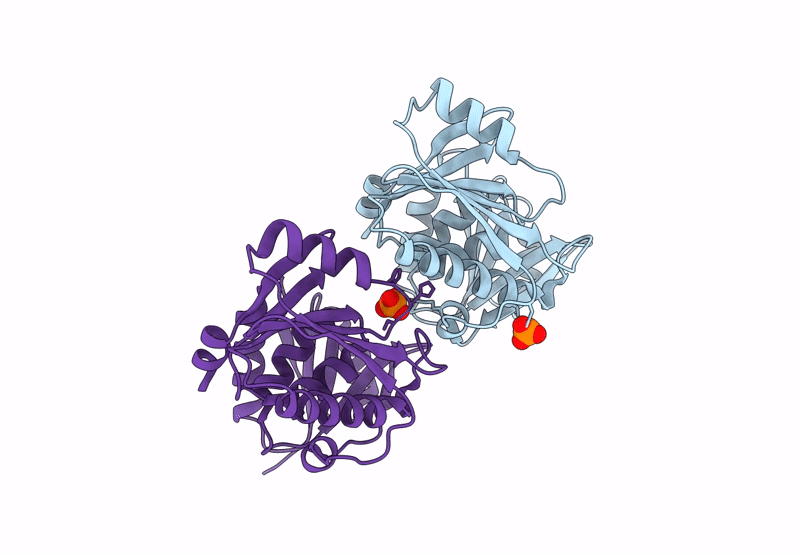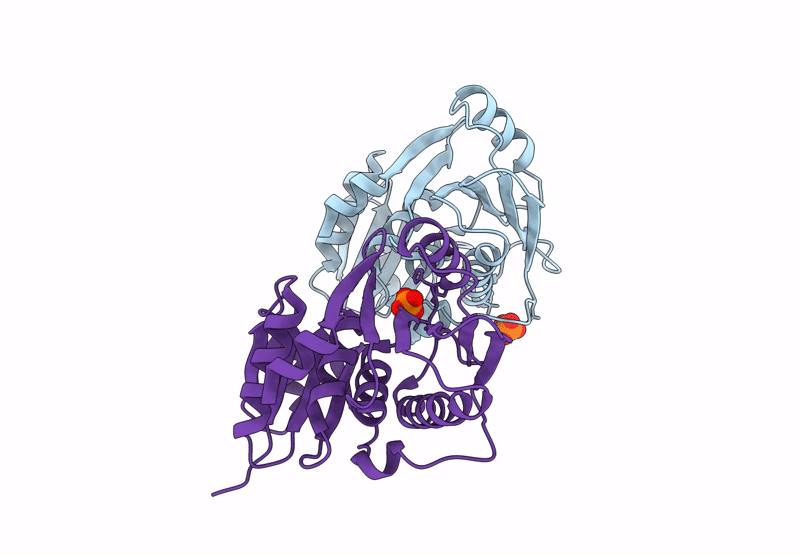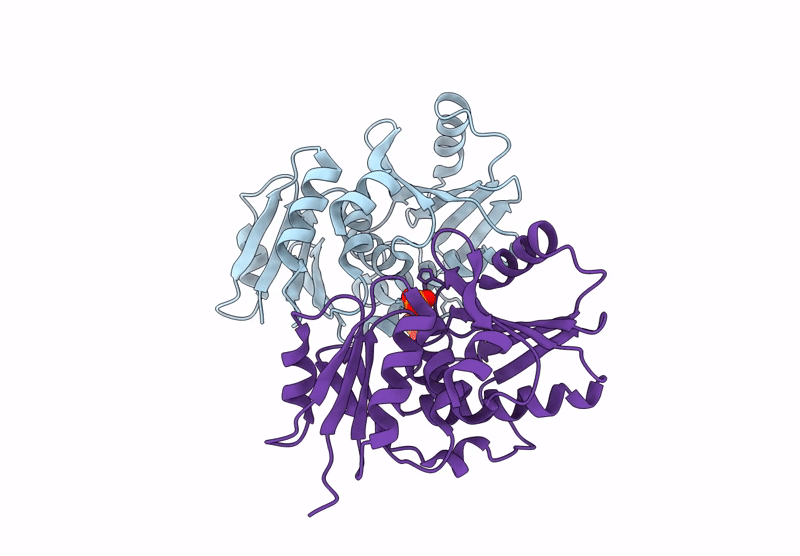
Structure And Catalytic Mechanism Of Sam-Amp Lyase In Clostridium Botulinum Cora-Associated Type Iii Crispr System
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: LYASE Ligands: A1IJB, MPD |
 |
Structure And Catalytic Mechanism Of Sam-Amp Lyase In Clostridium Botulinum Cora-Associated Type Iii Crispr System
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: LYASE Ligands: MPD |
 |
A Viral Saved Protein With Ring Nuclease Activity Subverts Type Iii Crispr Defence
Organism: Thermocrinis great boiling spring virus
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: VIRAL PROTEIN Ligands: PO4 |
 |
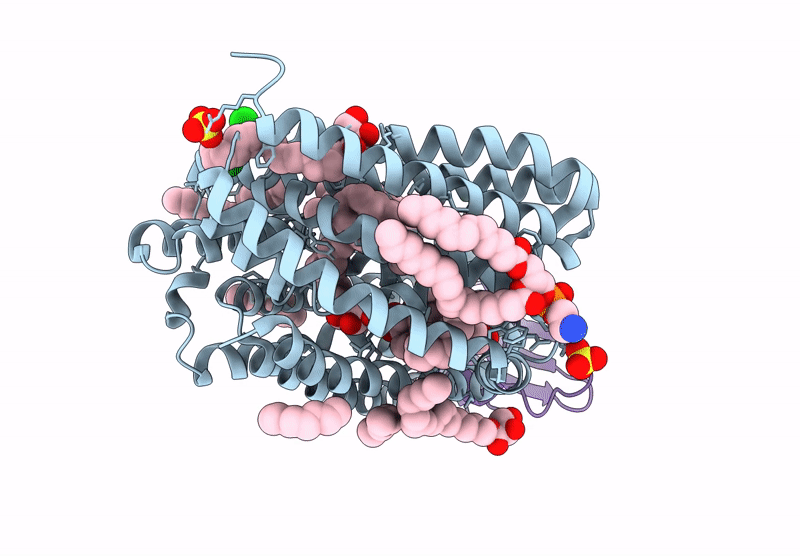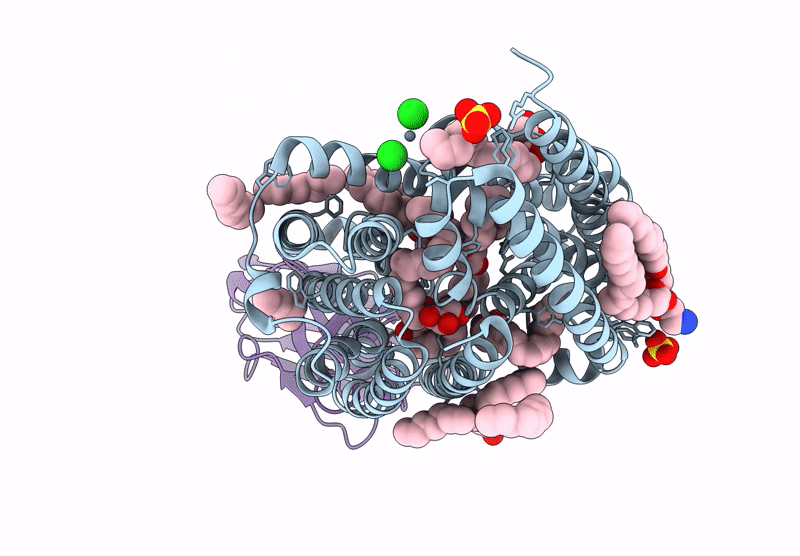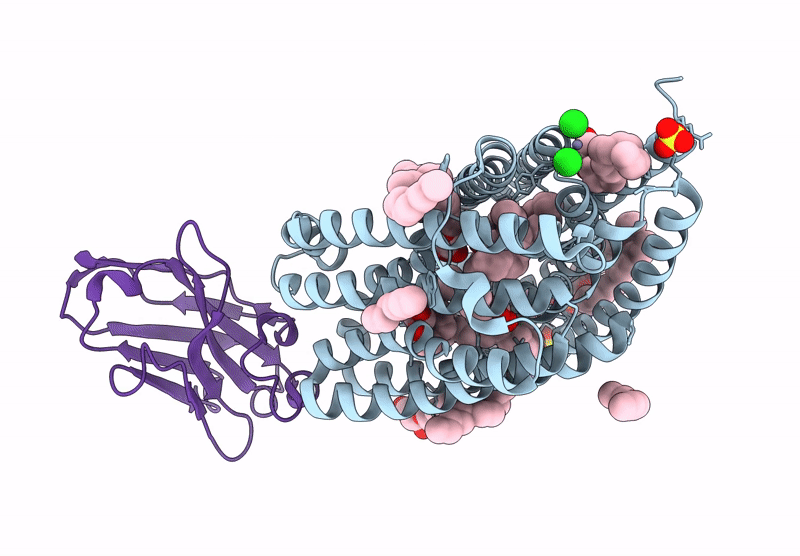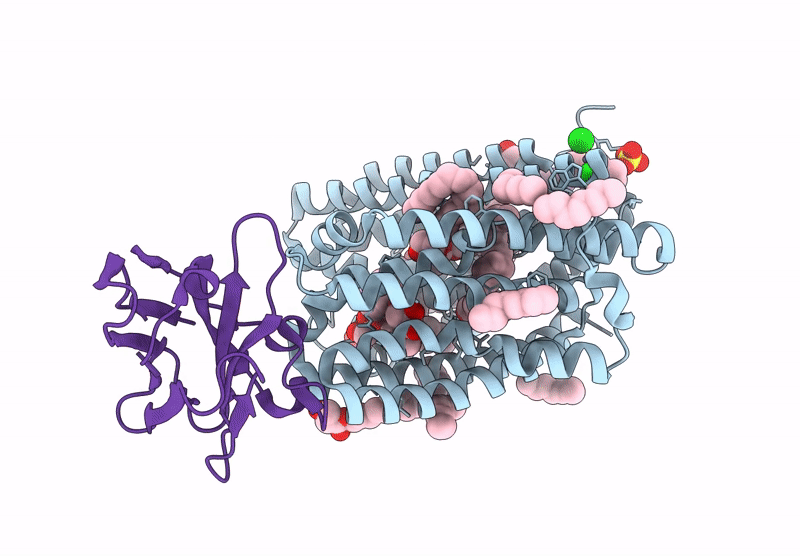
Lipid Iii Flippase Wzxe With Nb10 Nanobody In Outward-Facing Conformation At 2.7552 A
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN Ligands: MPG, OCT, SO4, ZN, CL |
 |
Lipid Iii Flippase Wzxe With Nb10 Nanobody In Outward-Facing Conformation At 0.9688 A
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN Ligands: MPG, LOP, OCT, CL, SO4, ZN |
 |
Lipid Iii Flippase Wzxe With Nb10 And Nb7 Nanobodies In Outward-Facing Conformation - Crystal 1
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN Ligands: PO4 |
 |
Lipid Iii Flippase Wzxe With Nb10 And Nb7 Nanobodies In Inward-Facing Conformation - Crystal 1
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN |
 |
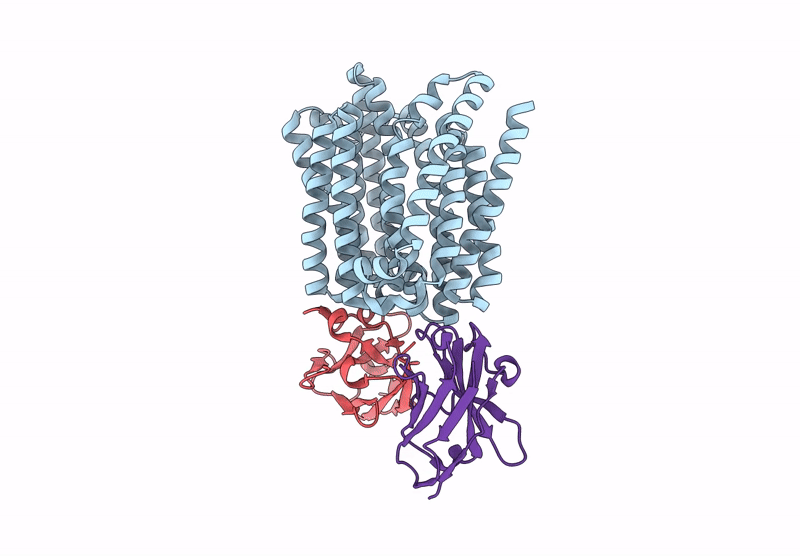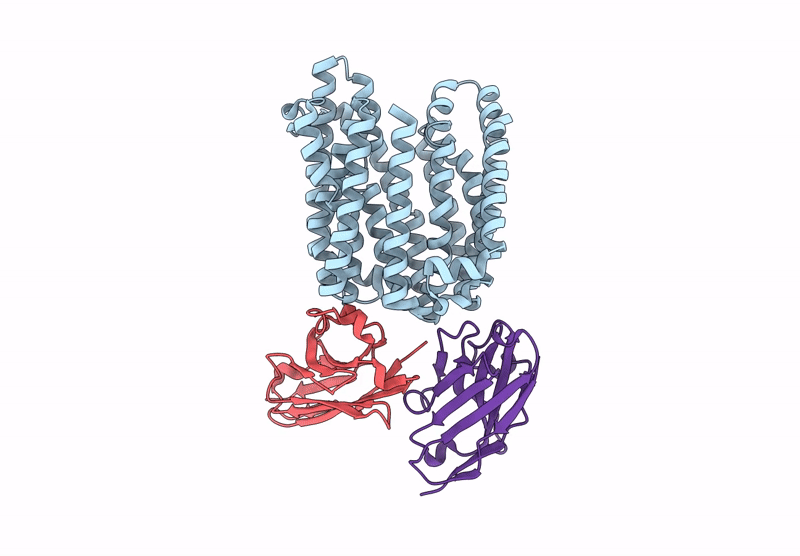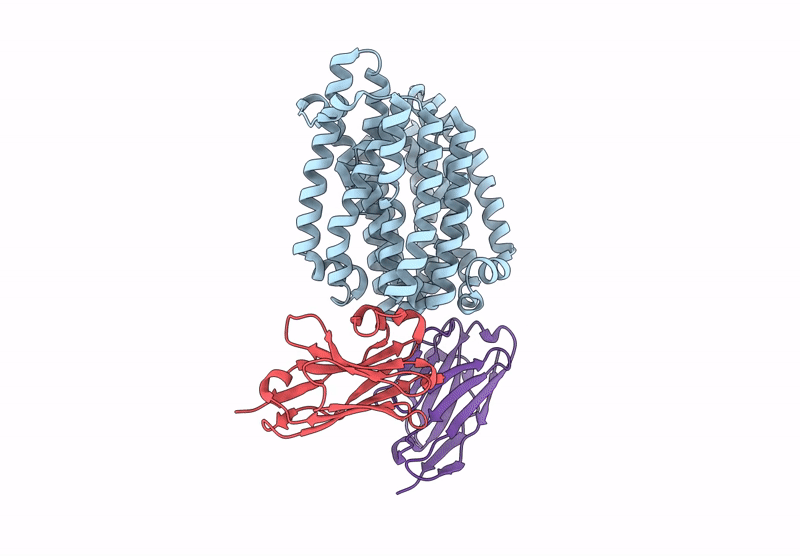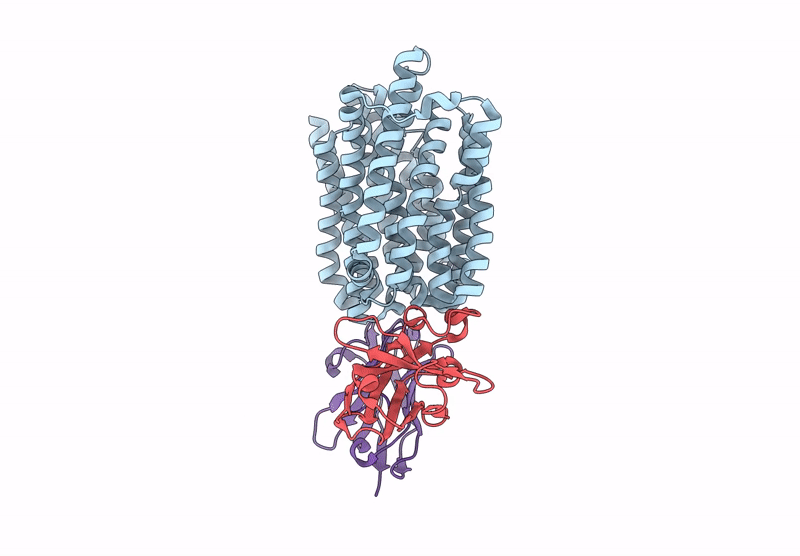
Lipid Iii Flippase Wzxe With Nb10 And Nb7 Nanobodies In Outward-Facing Conformation - Crystal 2
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN |
 |
Lipid Iii Flippase Wzxe With Nb10 And Nb7 Nanobodies In Inward-Facing Conformation - Crystal 2
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN |
 |
Organism: Streptococcus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.54 Å Release Date: 2023-10-04 Classification: RNA BINDING PROTEIN |
 |
Structure Of Csm6' From Streptococcus Thermophilus In Complex With Cyclic Hexa-Adenylate (Ca6)
Organism: Streptococcus thermophilus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2023-10-04 Classification: RNA BINDING PROTEIN |
 |
Organism: Aspergillus terreus (strain nih 2624 / fgsc a1156)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-10-20 Classification: CARBOHYDRATE |
 |
Structure Of Kdnase From Aspergillus Terrerus In Complex With 2-Keto-3-Deoxynononic Acid
Organism: Aspergillus terreus (strain nih 2624 / fgsc a1156)
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2021-10-20 Classification: CARBOHYDRATE Ligands: KDM, CA |
 |
Structure Of Kdnase From Aspergillus Terrerus In Complex With 2,3-Difluoro-2-Keto-3-Deoxynononic Acid
Organism: Aspergillus terreus (strain nih 2624 / fgsc a1156)
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2021-10-20 Classification: CARBOHYDRATE Ligands: FKD, K99, GOL, CL, CA |
 |
Structure Of Kdnase From Aspergillus Terrerus In Complex With 2,3-Didehydro-2,3-Dideoxy-D-Glycero-D-Galacto-Nonulosonic Acid.
Organism: Aspergillus terreus (strain nih 2624 / fgsc a1156)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-10-20 Classification: CARBOHYDRATE Ligands: KFN, GOL, CA |
 |
Structure Of Kdnase From Aspergillus Terrerus In Complex With 2-Keto-3-Deoxynononic Acid
Organism: Aspergillus terreus (strain nih 2624 / fgsc a1156)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-10-20 Classification: CARBOHYDRATE Ligands: GOL, KDM, CL |
 |
Structure Of Kdnase From Trichophyton Rubrum In Complex With 2-Keto-3-Deoxynononic Acid
Organism: Trichophyton rubrum
Method: X-RAY DIFFRACTION Resolution:0.91 Å Release Date: 2021-10-20 Classification: CARBOHYDRATE Ligands: KDM, GOL |
 |
Organism: Trichophyton rubrum
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-10-20 Classification: CARBOHYDRATE Ligands: PO4, NA, FKD |
 |
Structure Of Kdnase From Trichophyton Rubrum In Complex With 2,3-Didehydro-2,3-Dideoxy-D-Glycero-D-Galacto-Nonulosonic Acid.
Organism: Trichophyton rubrum
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2021-10-20 Classification: CARBOHYDRATE Ligands: KFN, NA |
 |
Structure Of Kdnase From Trichophyton Rubrum In Complex With 2-Keto-3-Deoxynononic Acid
Organism: Trichophyton rubrum
Method: X-RAY DIFFRACTION Resolution:0.99 Å Release Date: 2021-10-20 Classification: CARBOHYDRATE Ligands: KDM, GOL |

