Search Count: 49
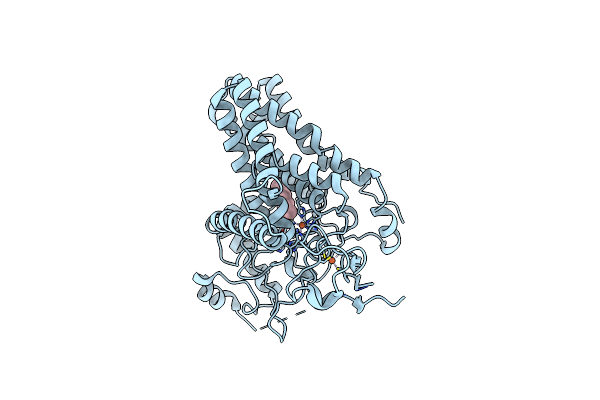 |
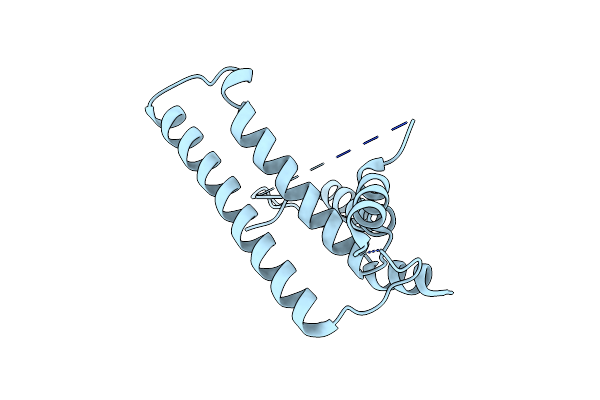
Cryo-Em Structure Of Alkane 1-Monooxygenase Alkb-Alkg Complex From Fontimonas Thermophila
Organism: Fontimonas thermophila
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: OXIDOREDUCTASE Ligands: FE, D12 |
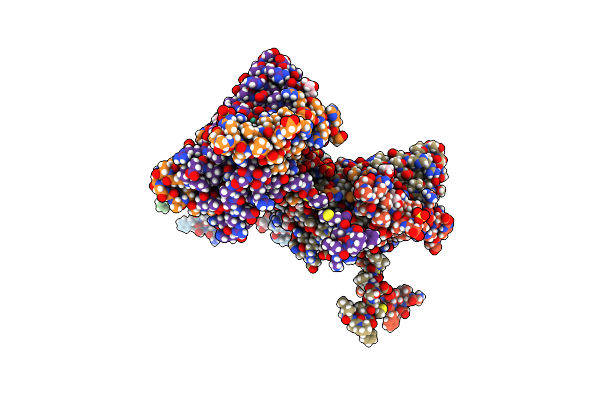 |
Structural And Functional Insights Of Dr2231 Protein, The Mazg-Like Nucleoside Triphosphate Pyrophosphohydrolase From Deinococcus Radiodurans, Complex With Gd
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-07-06 Classification: HYDROLASE Ligands: SO4, GOL, GD |
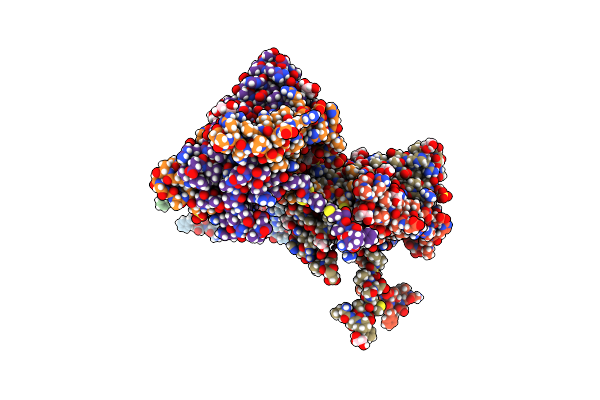 |
Crystal Structure Of Dr2231, The Mazg-Like Protein From Deinococcus Radiodurans, Complex With Manganese
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-07-06 Classification: HYDROLASE Ligands: MN, SO4, GOL |
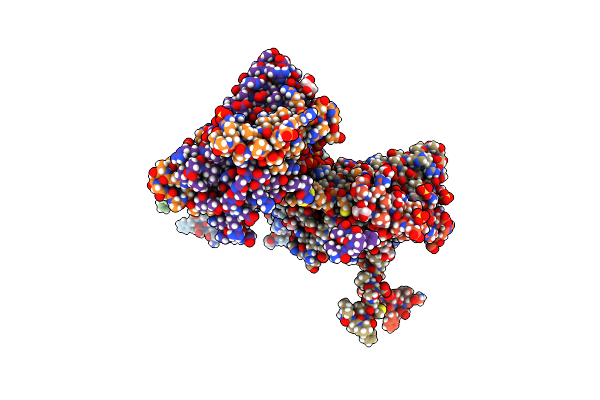 |
Crystal Structure Of Dr2231, The Mazg-Like Protein From Deinococcus Radiodurans, Apo Structure
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-07-06 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
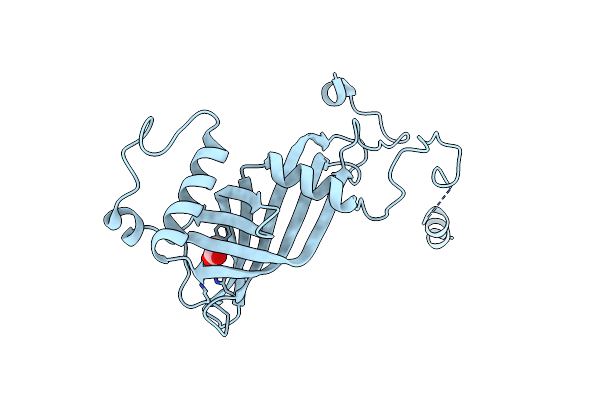
Structural And Functional Insights Of Dr2231 Protein, The Mazg-Like Nucleoside Triphosphate Pyrophosphohydrolase From Deinococcus Radiodurans, Native Form
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-07-06 Classification: HYDROLASE Ligands: CL |
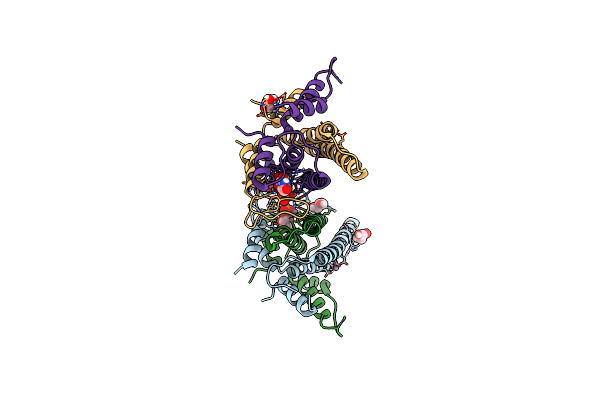 |
Structural And Functional Insights Of Dr2231 Protein, The Mazg-Like Nucleoside Triphosphate Pyrophosphohydrolase From Deinococcus Radiodurans, Complexed With Mn And Dump
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2011-07-06 Classification: HYDROLASE Ligands: UMP, MN, GOL |
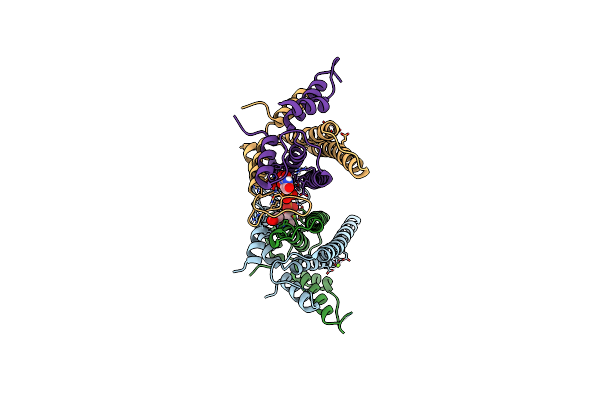 |
Structural And Functional Insights Of Dr2231 Protein, The Mazg-Like Nucleoside Triphosphate Pyrophosphohydrolase From Deinococcus Radiodurans, Complexed With Mg And Dump
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2011-07-06 Classification: HYDROLASE Ligands: MG, UMP, CL, ACT, GOL |
 |
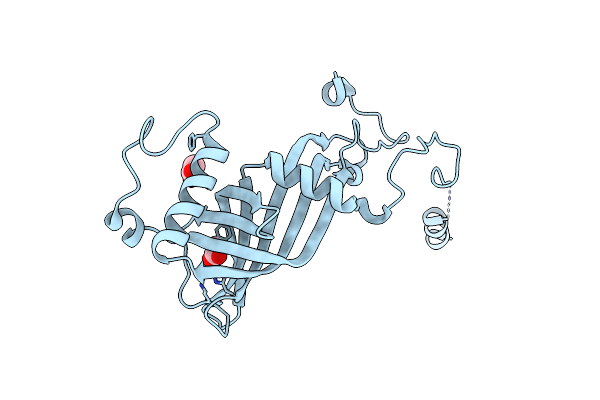
H71A Mutant Of The Antibiotic Resistance Protein Nima From Deinococcus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-11-17 Classification: OXIDOREDUCTASE Ligands: ACT |
 |
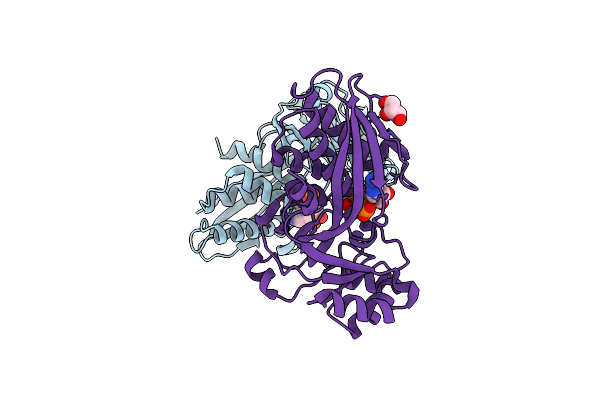
H71S Mutant Of The Antibiotic Resistance Protein Nima From Deinococcus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2010-11-17 Classification: OXIDOREDUCTASE Ligands: ACT |
 |
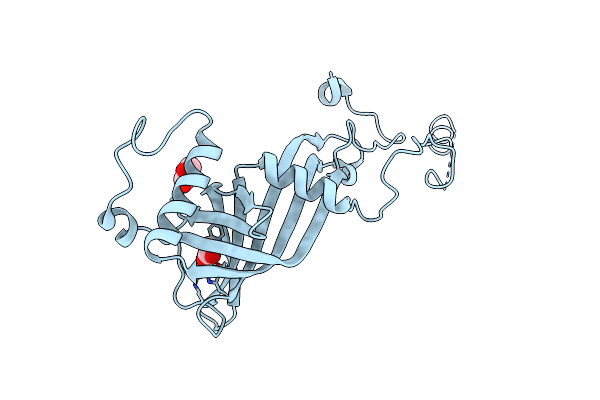
A Triclinic Crystal Form Of E. Coli 4-Diphosphocytidyl-2C-Methyl-D- Erythritol Kinase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-03-09 Classification: TRANSFERASE Ligands: ADP, GOL |
 |
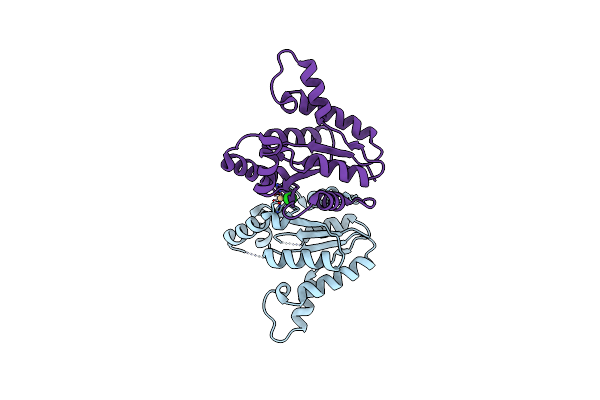
High Resolution Crystal Structure Of The Antibiotic Resistance Protein Nima From Deinococcus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2008-08-19 Classification: OXIDOREDUCTASE Ligands: ACT, PYR |
 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2008-06-17 Classification: TRANSFERASE Ligands: CD, CL |
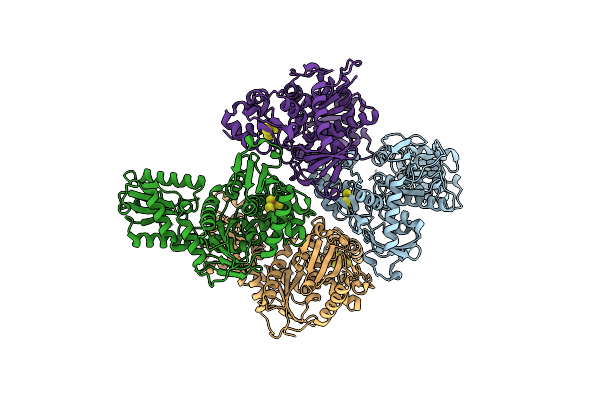 |
The Crystal Structure Of Dr2241 From Deinococcus Radiodurans At 1.9 A Resolution Reveals A Multi-Domain Protein With Structural Similarity To Chelatases But Also With Two Additional Novel Domains
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-06-19 Classification: RIBOSOMAL PROTEIN Ligands: SF4 |
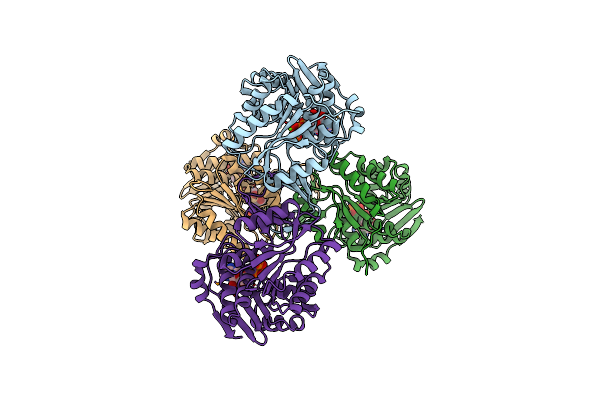 |
Structure Of Staphylococcus Aureus D-Tagatose-6-Phosphate Kinase With Cofactor And Substrate
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-04-24 Classification: TRANSFERASE Ligands: MG, ANP, TA6 |
 |
Structure Of Staphylococcus Aureus D-Tagatose-6-Phosphate Kinase In Complex With Adp
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-04-24 Classification: TRANSFERASE Ligands: ADP |
 |
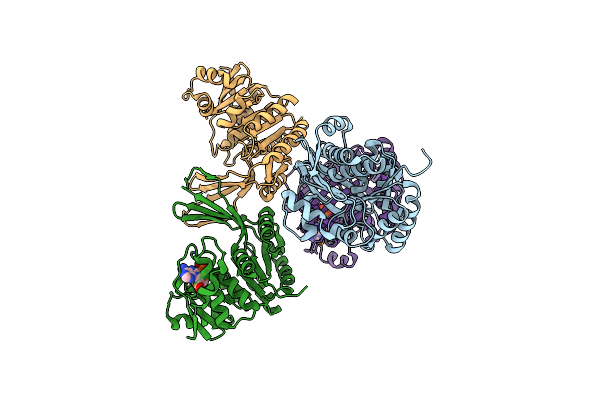
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2007-02-20 Classification: DNA BINDING PROTEIN Ligands: FE, MG |
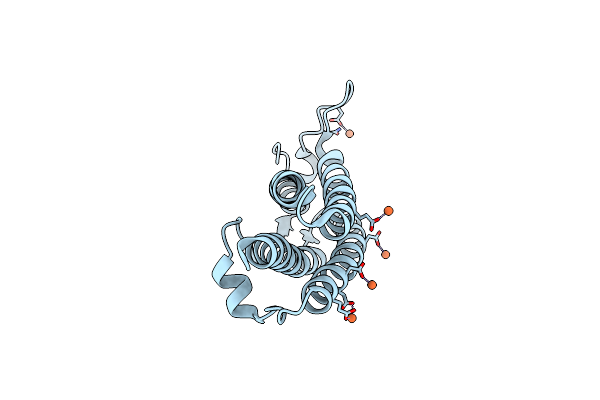 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-02-20 Classification: DNA BINDING PROTEIN Ligands: FE, CL |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2006-07-17 Classification: TRANSCRIPTION Ligands: FMT, GOL |
 |
Nikr From Helicobacter Pylori In Closed Trans-Conformation And Nickel Bound To 2F, 2X And 2I Sites.
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-07-17 Classification: TRANSCRIPTIONAL REGULATION Ligands: FMT, GOL, NI, CIT |
 |
Nikr From Helicobacter Pylori In Closed Trans-Conformation And Nickel Bound To 4 Intermediary Sites
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2006-07-17 Classification: TRANSCRIPTIONAL REGULATION Ligands: GOL, CL, NI |

