Search Count: 15
 |
Burkholderia Pseudomallei Disulfide Bond Forming Protein A (Dsba) Liganded With Fragment Bromophenoxy Propanamide
Organism: Burkholderia pseudomallei (strain k96243)
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-01-05 Classification: OXIDOREDUCTASE Ligands: YCS |
 |
Burkholderia Pseudomallei Disulfide Bond Forming Protein A (Dsba) Liganded With Fragment 4-Methoxy-N-Phenylbenzenesulfonamide
Organism: Burkholderia pseudomallei (strain k96243)
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2022-01-05 Classification: OXIDOREDUCTASE Ligands: YCY, SO4 |
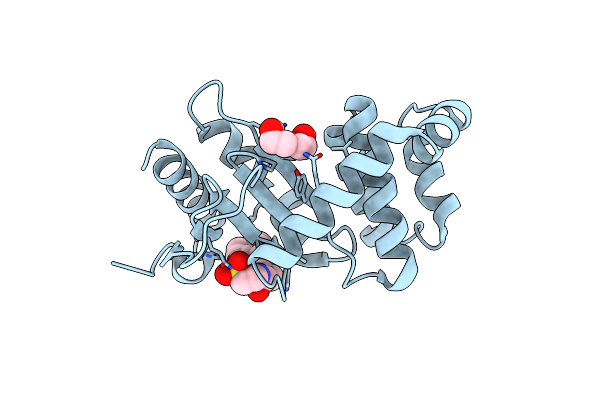 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2021-12-29 Classification: OXIDOREDUCTASE Ligands: PEG, TCH |
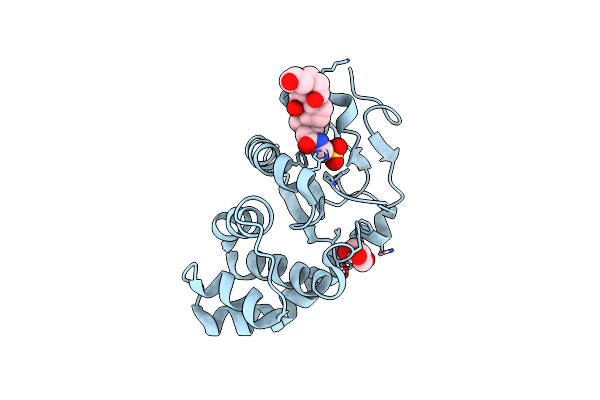 |
Crystal Structure Of Vibrio Cholerae Dsba In Complex With Bile Salt Taurocholate
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2021-12-29 Classification: OXIDOREDUCTASE Ligands: GOL, TCH |
 |
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-01-06 Classification: ANTIMICROBIAL PROTEIN Ligands: MLI |
 |
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2018-03-21 Classification: OXIDOREDUCTASE |
 |
Model Structure Of The Oxidized Padsba1 And 3-[(2-Methylbenzyl)Sulfanyl]-4H-1,2,4-Triazol-4-Amine Complex
Organism: Pseudomonas aeruginosa
Method: SOLUTION NMR Release Date: 2017-04-12 Classification: OXIDOREDUCTASE Ligands: 1YO |
 |
Organism: Chlamydia trachomatis serovar a
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2017-01-11 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Pseudomonas Aeruginosa Dsba E82I In Complex With Mips-0000851 (3-[(2-Methylbenzyl)Sulfanyl]-4H-1,2,4-Triazol-4-Amine)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2016-10-05 Classification: OXIDOREDUCTASE Ligands: 1YO, GOL, MES |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2015-12-09 Classification: OXIDOREDUCTASE Ligands: P6G |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-12-09 Classification: OXIDOREDUCTASE Ligands: MES, GOL |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-12-09 Classification: OXIDOREDUCTASE Ligands: MES, EDO, PG4 |
 |
Organism: Pseudomonas aeruginosa
Method: SOLUTION NMR Release Date: 2014-11-12 Classification: OXIDOREDUCTASE |
 |
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-08-14 Classification: OXIDOREDUCTASE Ligands: GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-04-27 Classification: IMMUNE SYSTEM |

