Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Crystal Structure Of Candida Glabrata Bdf1 Bromodomain 1 In The Unbound State
Organism: Nakaseomyces glabratus
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2024-10-30 Classification: TRANSCRIPTION |
 |
Crystal Structure Of Candida Glabrata Bdf1 Bromodomain 1 Bound To A Pyrazole Ligand
Organism: Nakaseomyces glabratus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2024-10-30 Classification: TRANSCRIPTION Ligands: Y7F, SO4 |
 |
Crystal Structure Of Candida Glabrata Bdf1 Bromodomain 1 Bound To A Phenyltriazine Ligand
Organism: Nakaseomyces glabratus
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2024-10-30 Classification: TRANSCRIPTION Ligands: Y78, PO4, K |
 |
Crystal Structure Of Candida Glabrata Bdf1 Bromodomain 2 In The Unbound State
Organism: Nakaseomyces glabratus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2024-10-30 Classification: TRANSCRIPTION Ligands: MPD, GOL, ACT |
 |
Organism: Nakaseomyces glabratus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-10-30 Classification: TRANSCRIPTION Ligands: GOL, SO4, 1GH |
 |
Crystal Structure Of Candida Glabrata Bdf1 Bromodomain 2 Bound To A Pyridoindole Ligand
Organism: Nakaseomyces glabratus
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2024-10-30 Classification: TRANSCRIPTION Ligands: Y7B |
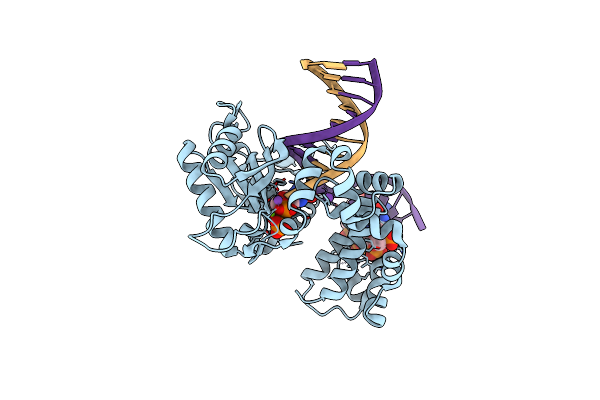 |
Ternary Complex Crystal Structure Of K289M Variant Of Dna Polymerase Beta With Dgtp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2020-01-08 Classification: transcription/dna Ligands: MG, NA, CL, DGT |
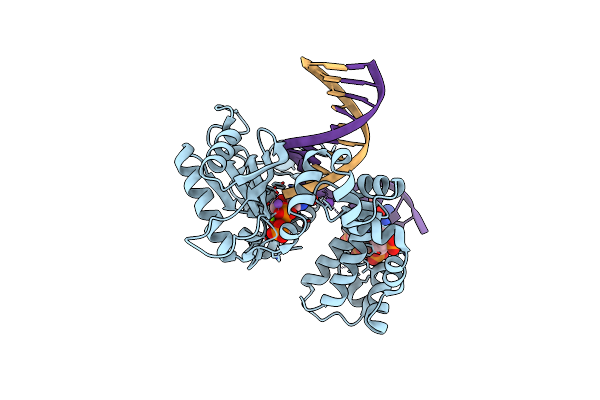 |
Ternary Complex Crystal Structure Of K289M Variant Of Dna Polymerase Beta With Beta-Gamma Chf Analog Of Dgtp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2020-01-08 Classification: transcription/dna Ligands: GFH, MG, NA, CL |
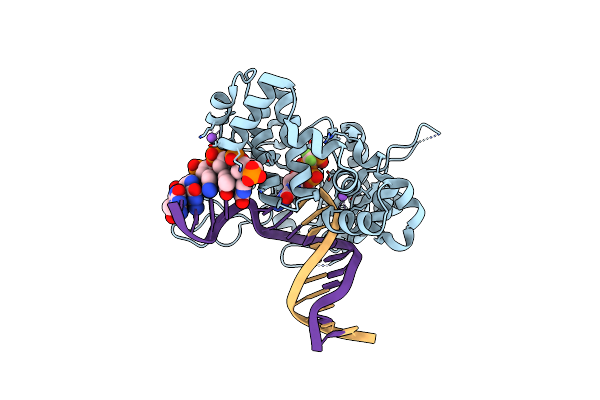 |
Ternary Complex Crystal Structure Of K289M Variant Of Dna Polymerase Beta With Beta-Gamma Difluoro Analogue Of Dgtp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-01-08 Classification: transcription/dna |
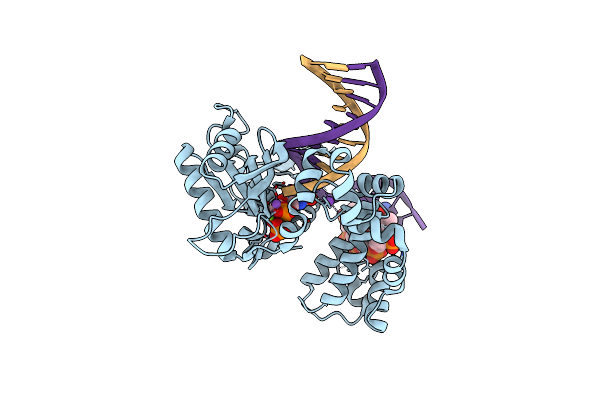 |
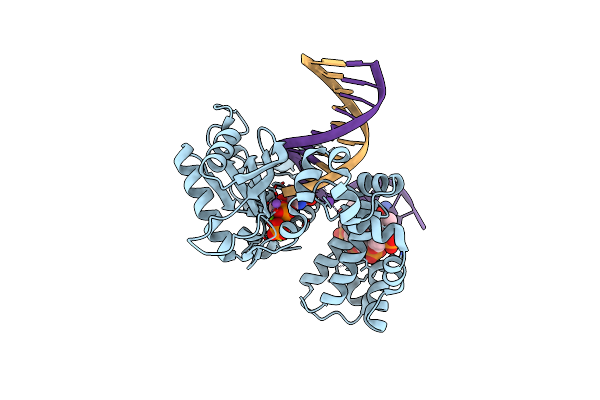
Ternary Complex Crystal Structure Of Dna Polymerase Beta With "Hot-Spot Sequence" With Dgtp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-01-08 Classification: transcription/dna Ligands: MG, NA, CL, DGT |
 |
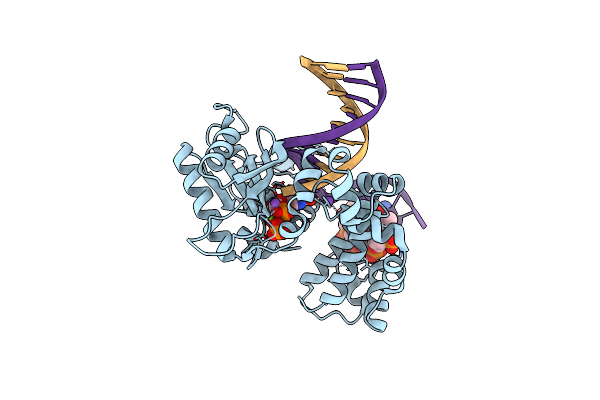
Ternary Complex Crystal Structure Of Dna Polymerase Beta With "Hot-Spot Sequence" With Beta-Gamma Chf Analogue Of Dgtp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-01-08 Classification: transcription/dna Ligands: MG, NA, CL, GFH |
 |
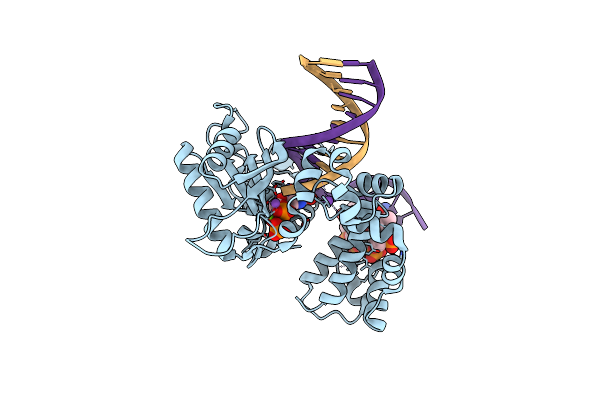
Ternary Complex Crystal Structure Of Dna Polymerase Beta With "Hot-Spot Sequence" With Beta-Gamma-Methylene Dgtp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2020-01-08 Classification: transcription/dna Ligands: MG, NA, CL, GGH |
 |
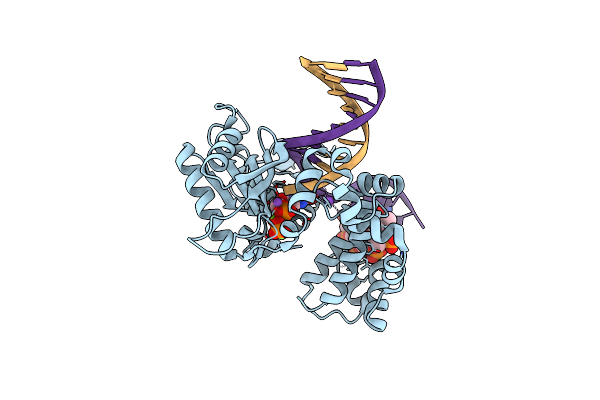
Ternary Complex Crystal Structure Of K289M Variant Of Dna Polymerase Beta With "Hot-Spot Sequence" With Dgtp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2020-01-08 Classification: transcription/dna |
 |
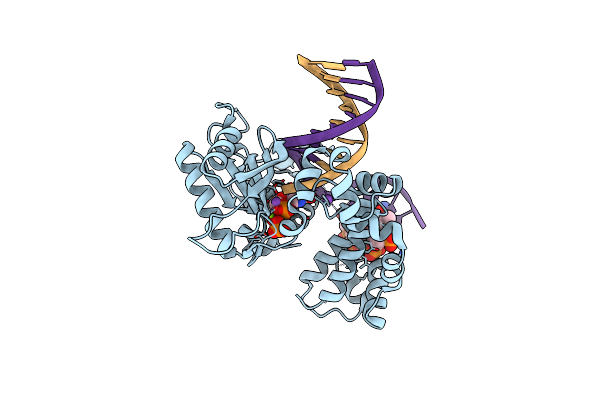
Ternary Complex Crystal Structure Of K289M Variant Of Dna Polymerase Beta With "Hot-Spot Sequence" With Beta-Gamma Chf Analogue Of Dgtp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2020-01-08 Classification: transcription/dna Ligands: MG, NA, CL, GFH |
 |
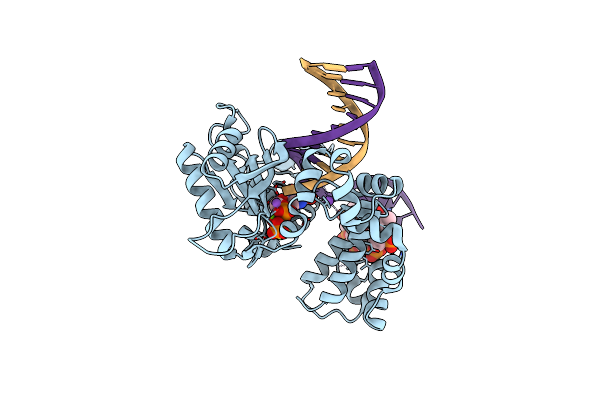
Ternary Complex Crystal Structure Of K289M Variant Of Dna Polymerase Beta With "Hot-Spot Sequence" With Beta-Gamma Methylene Dgtp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2020-01-08 Classification: transcription/dna |
 |
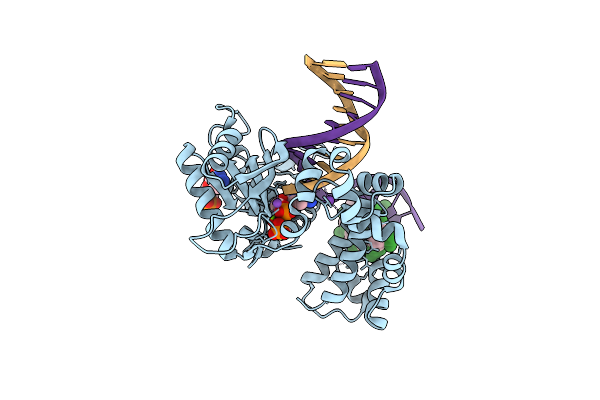
Ternary Complex Crystal Structure Of K289M Variant Of Dna Polymerase Beta With "Hot-Spot Sequence" With Beta-Gamma Cf2 Analogue Of Dgtp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2020-01-08 Classification: transcription/dna Ligands: MG, NA, GFF, CL |
 |
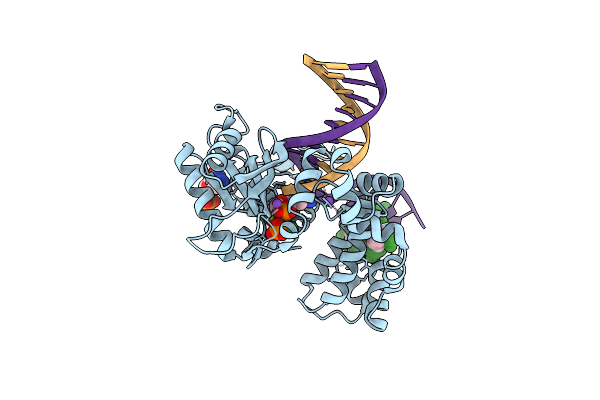
Ternary Complex Crystal Structure Of Dna Polymerase Beta With R-Isomer Of Beta-Gamma-Chf-Dctp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-10-31 Classification: TRANSFERASE/DNA Ligands: MG, NA, CL, F3C, DC |
 |
Ternary Complex Crystal Structure Of Dna Polymerase Beta With S-Isomer Of Beta-Gamma-Chcl-Dctp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2018-10-31 Classification: TRANSFERASE/DNA Ligands: MG, NA, CL, DJJ, DC |
 |
Ternary Complex Crystal Structure Of Dna Polymerase Beta With A Dideoxy Terminated Primer With Ch-Ch3, Beta, Gamma Datp Analogue
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-08-22 Classification: TRANSFERASE/DNA |
 |
Ternary Complex Crystal Structure Of Dna Polymerase Beta With A Dideoxy Terminated Primer With Ccl2, Beta, Gamma Datp Analogue
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-07-25 Classification: TRANSFERASE/DNA Ligands: VA7, MG, NA |