Search Count: 22
 |
Human Nucleosome Structure On Nickel-Nta Lipid Affinity Grid (C2 Refinement)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: DNA BINDING PROTEIN |
 |
Human Nucleosome Structure On Nickel-Nta Lipid Affinity Grid (C1 Refinement)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: DNA BINDING PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: EXOCYTOSIS |
 |
Co-Crystal Structure Of Maltose Binding Protein (Mbp)-Human Senp3 Fusion Protein In Complex With Pelp1 Peptide
|
 |
Organism: Homo sapiens, Synthetic construct, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: GENE REGULATION/DNA Ligands: FE, ZN, OH0 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.90 Å Release Date: 2025-04-16 Classification: CELL CYCLE Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.90 Å Release Date: 2025-04-16 Classification: CELL CYCLE Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: NUCLEAR PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-10-02 Classification: TRANSFERASE Ligands: A1AZ6, EDO, UNX |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-10-02 Classification: TRANSFERASE Ligands: A1AZ5, SO4, EDO, UNX |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-02-22 Classification: GENE REGULATION/DNA Ligands: OH0, FE |
 |
Crystal Structure Of Kdm2A Histone Demethylase Catalytic Domain In Complex With An H3C36 Peptide Modified By Unc8015
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2023-02-22 Classification: GENE REGULATION Ligands: FE, OH0 |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-05-04 Classification: STRUCTURAL PROTEIN/DNA/TRANSFERASE |
 |
Crystal Structure Of Human Mpp8 Chromodomain In Complex With Peptidomimetic Ligand Unc5246
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2021-09-01 Classification: GENE REGULATION |
 |
Organism: Homo sapiens, Synthetic construct, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2020-09-16 Classification: DNA Binding Protein/DNA/Transferase Ligands: ZN |
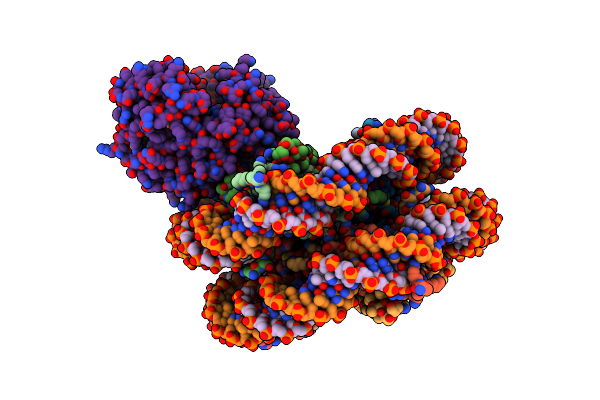 |
Organism: Homo sapiens, Synthetic construct, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2020-09-16 Classification: Dna Binding Protein/DNA/Transferase Ligands: ZN |
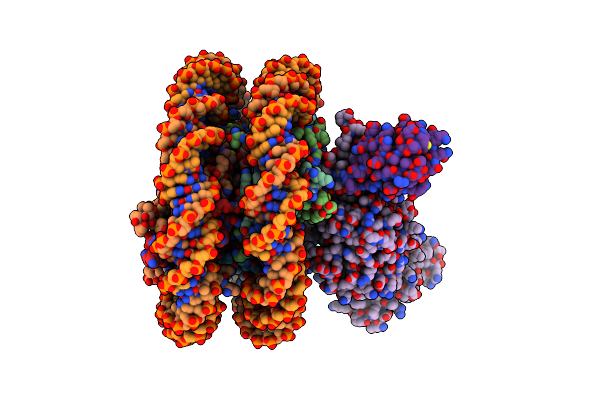 |
Organism: Xenopus laevis, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2019-02-13 Classification: GENE REGULATION |
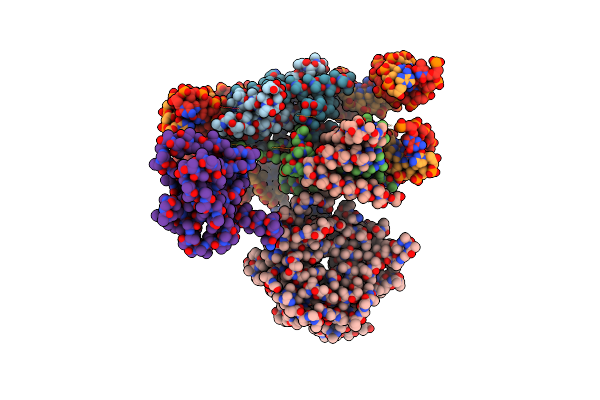 |
Structural Model Of Set8 Histone H4 Lys20 Methyltransferase Bound To Nucleosome Core Particle
Organism: Xenopus laevis, Synthetic construct, Saccharomyces cerevisiae (strain atcc 204508 / s288c), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.50 Å Release Date: 2016-03-23 Classification: Transferase/DNA |
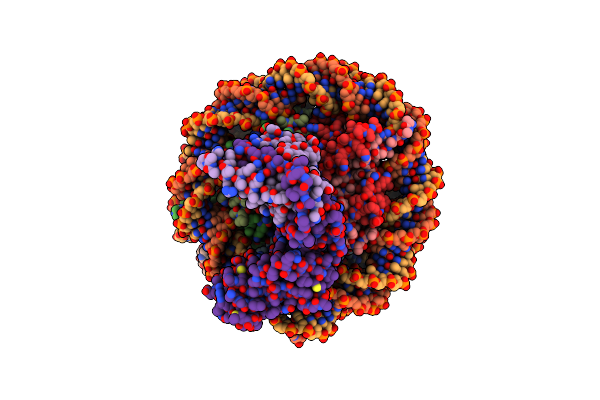 |
Crystal Structure Of The Ring1B/Bmi1/Ubch5C Prc1 Ubiquitylation Module Bound To The Nucleosome Core Particle
Organism: Xenopus laevis, Synthetic dna, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.28 Å Release Date: 2014-11-05 Classification: Structural Protein/DNA Ligands: ZN |
 |
Organism: Homo sapiens, Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-06-01 Classification: TRANSCRIPTION/NUCLEAR PROTEIN |

