Search Count: 47
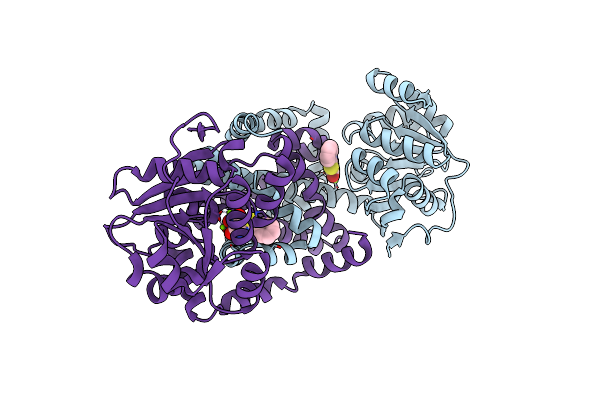 |
Crystal Structure Of Staphylococcus Aureus Ketol-Acid Reductoisomerase In Complex With Mg2+, And Jk-5-114
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-04-09 Classification: ISOMERASE Ligands: A1AKJ, MG |
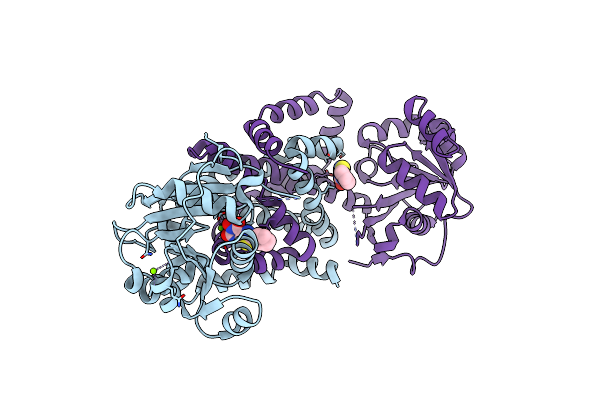 |
Crystal Structure Of Staphylococcus Aureus Ketol-Acid Reductoisomerase In Complex With Mg2+, And Jk-5-115
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-04-09 Classification: ISOMERASE Ligands: MG, A1AKY |
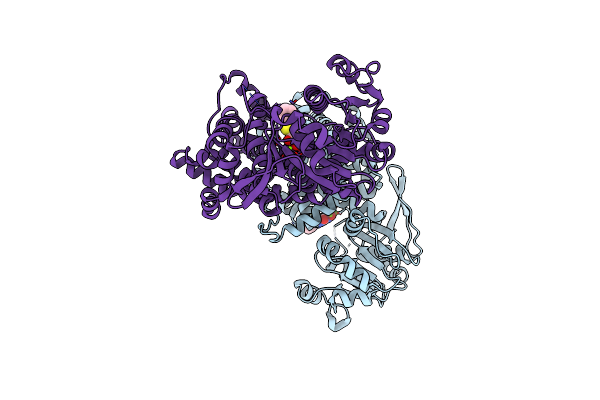 |
Crystal Structure Of Oryza Sativa Ketol-Acid Reductoisomerase In Complex With Mg2+, And Jk-5-114
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Release Date: 2025-04-09 Classification: ISOMERASE Ligands: MG, A1AKJ |
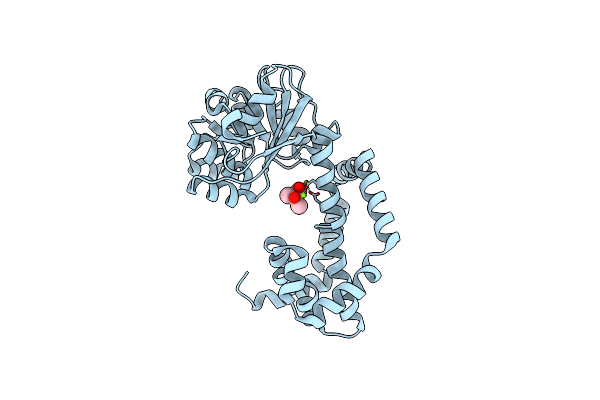 |
Crystal Structure Of Campylobacter Jejuni Ketol-Acid Reductoisomerase In Complex With 2-Acetolactate
Organism: Campylobacter jejuni subsp. jejuni
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-04-24 Classification: OXIDOREDUCTASE Ligands: MG, X2X, CL |
 |
Campylobacter Jejuni Keto-Acid Reductoisomerase In Complex With Intermediate And Nadp+
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2024-04-24 Classification: OXIDOREDUCTASE Ligands: WXU, MG, NCA, NDP, CL |
 |
Campylobacter Jejuni Ketol-Acid Reductoisomerase In Complex With Nadp+ And Hmkb
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2024-04-24 Classification: OXIDOREDUCTASE Ligands: MG, CL, NDP, WXU |
 |
Campylobacter Jejuni Ketol-Acid Reductoisomerase In Complex With Nadph And Hoe704
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2024-04-24 Classification: OXIDOREDUCTASE Ligands: MG, NDP, X9W |
 |
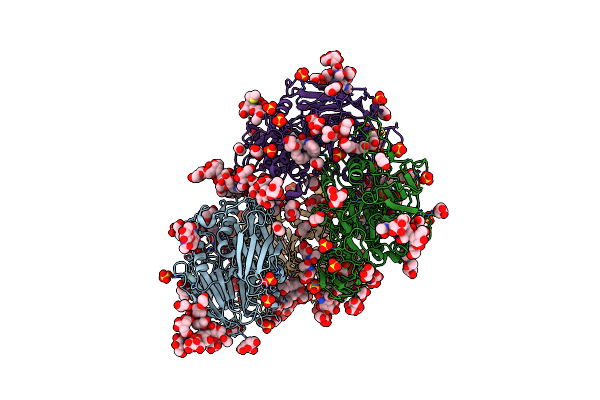
Campylobacter Jejuni Ketol-Acid Reductoisomerase In Complex With 2,3-Dihydroxy-3-Isovalerate.
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2024-04-24 Classification: OXIDOREDUCTASE Ligands: MG, XAI |
 |
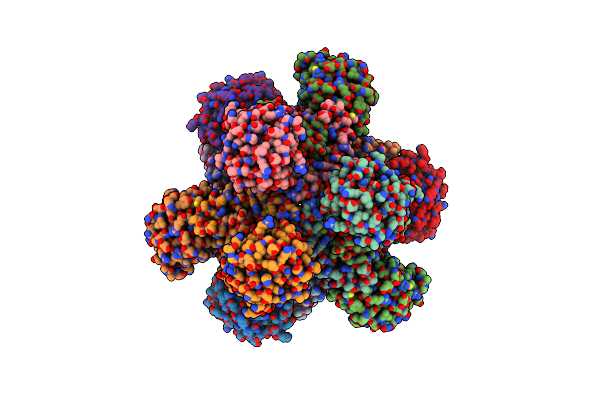
Crystal Structure Of Red Kidney Bean Purple Acid Phosphatase In Complex With An Alpha-Aminonaphthylmethylphosphonic Acid Inhibitor
Organism: Phaseolus vulgaris
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-08-02 Classification: METAL BINDING PROTEIN Ligands: ZN, FE, FLC, PGE, NAG, SO4, EDO, GOL, CL, R9X, NA, DMS |
 |
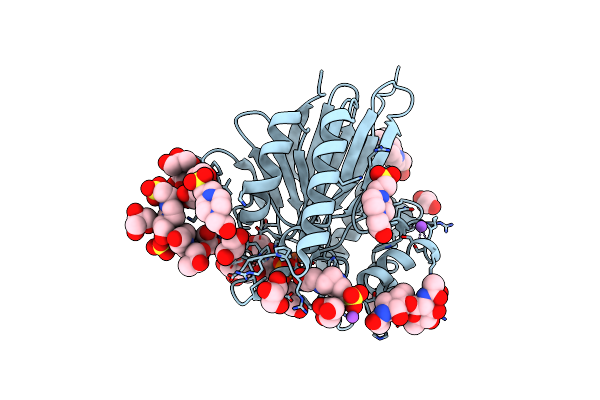
Apo Form Cryo-Em Structure Of Campylobacter Jejune Ketol-Acid Reductoisommerase Crosslinked By Glutaraldehyde
Organism: Campylobacter jejuni
Method: ELECTRON MICROSCOPY Release Date: 2023-02-01 Classification: ISOMERASE Ligands: PTD |
 |
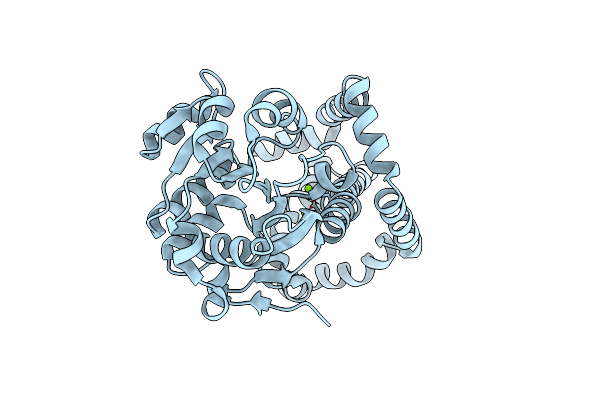
Crystal Structure Of Pig Purple Acid Phosphatase In Complex With 4-(2-Hydroxyethyl)-1-Piperazineethanesulfonic Acid (Hepes) And Glycerol
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-03-23 Classification: HYDROLASE Ligands: EPE, GOL, FE, PO4, EDO, PGE, NA |
 |
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2022-01-12 Classification: ISOMERASE Ligands: MG |
 |
Crystal Structure Of Pig Purple Acid Phosphatase In Complex With L-Glutamine, (Poly)Ethylene Glycol Fragments And Glycerol
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-01-12 Classification: HYDROLASE Ligands: 1PE, P6G, PG4, GLN, PO4, EDO, PGE, GOL, CIT, NA, FE |
 |
Crystal Structure Of Pig Purple Acid Phosphatase In Complex With Maybridge Fragment Cc063346, Dimethyl Sulfoxide And Citrate
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-01-12 Classification: HYDROLASE Ligands: DMS, PGE, R2J, NAG, PO4, CIT, GOL, FE, EDO, NA, CL |
 |
Crystal Structure Of Staphylococcus Aureus Ketol-Acid Reductoisomerase In Complex With Mg2+ And Nsc116565
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2021-07-14 Classification: ISOMERASE, Oxidoreductase Ligands: MG, WBY |
 |
Crystal Structure Of Staphylococcus Aureus Ketol-Acid Reductoisomerase In Complex With Mg2+, Nadph, And Nsc116565
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2021-07-14 Classification: ISOMERASE/Inhibitor Ligands: NAP, NDP, MG, WBY |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2020-07-15 Classification: TRANSFERASE Ligands: TPP, MG, FAD, 60G, ATP |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2020-07-15 Classification: PLANT PROTEIN Ligands: MG, FAD, TPP |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2020-07-15 Classification: PLANT PROTEIN Ligands: MG, FAD, TPP, VAL |
 |
Hybrid Acetohydroxyacid Synthase Complex Structure With Cryptococcus Neoformans Ahas Catalytic Subunit And Saccharomyces Cerevisiae Ahas Regulatory Subunit
Organism: Cryptococcus neoformans, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2020-07-15 Classification: TRANSFERASE Ligands: FAD, 8GF, DPO, MG, VAL |

