Search Count: 18
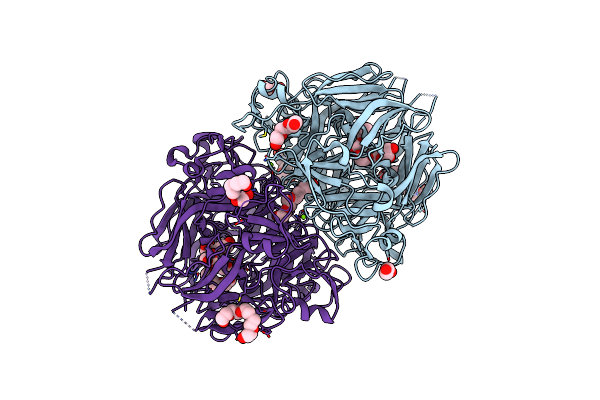 |
Adaptive Mechanism Of Collagen Iv Scaffold Assembly In Drosophila: Crystal Structure Of Recombinant Nc1 Hexamer
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-11-08 Classification: STRUCTURAL PROTEIN Ligands: PGE, CL, MG, PG4, PEG, EDO, 1PE, PO4 |
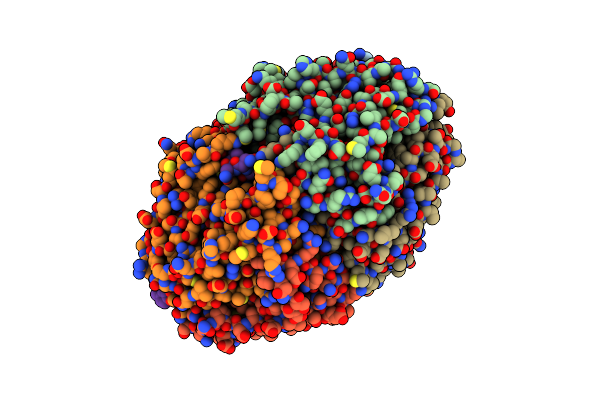 |
Adaptive Mechanism Of Collagen Iv Scaffold Assembly In Drosophila: Crystal Structure Of Tissue-Extracted Nc1 Hexamer
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2023-11-08 Classification: STRUCTURAL PROTEIN Ligands: CL, CA |
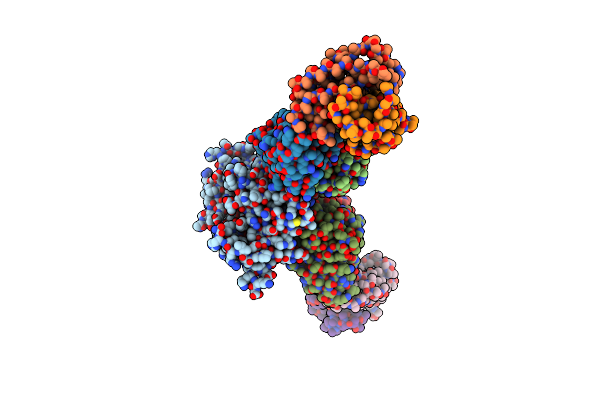 |
Cryoem Structure Of Hantavirus Andv Gn(H) Protein Complex With 2Fabs Andv-5 And Andv-34
Organism: Andes orthohantavirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-01-27 Classification: TRANSCRIPTION, TRANSFERASE Ligands: FLC |
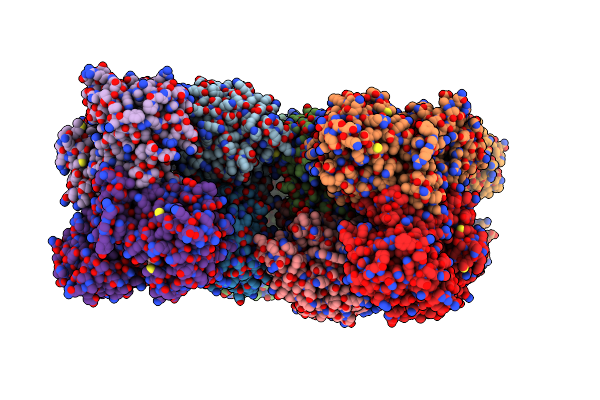 |
Caenorhabditis Elegans Udp-Glucose Dehydrogenase In Complex With Udp-Xylose
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2019-05-22 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: UDX |
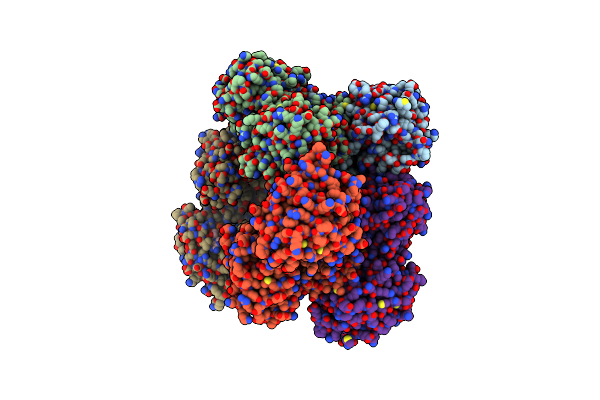 |
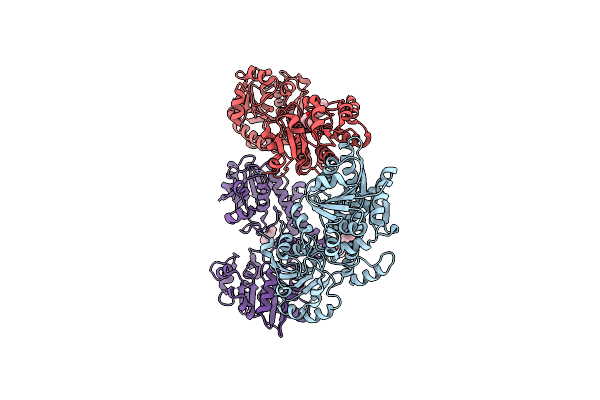
Human Udp-Glucose Dehydrogenase With Udp-Xylose Bound To The Co-Enzyme Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-07-19 Classification: OXIDOREDUCTASE Ligands: UDX, ADP |
 |
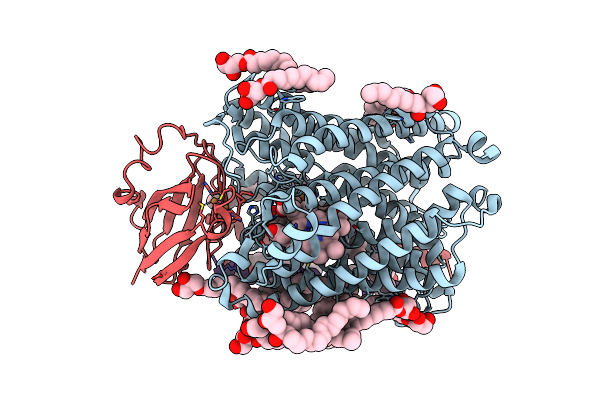
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2017-07-19 Classification: OXIDOREDUCTASE Ligands: PG5 |
 |
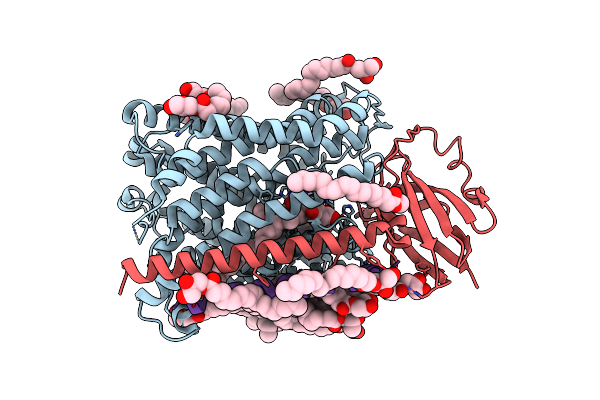
Structure Of Recombinant Cytochrome Ba3 Oxidase Mutant Y133F From Thermus Thermophilus
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-01-16 Classification: OXIDOREDUCTASE Ligands: CU, HEM, HAS, PER, OLC, CUA |
 |
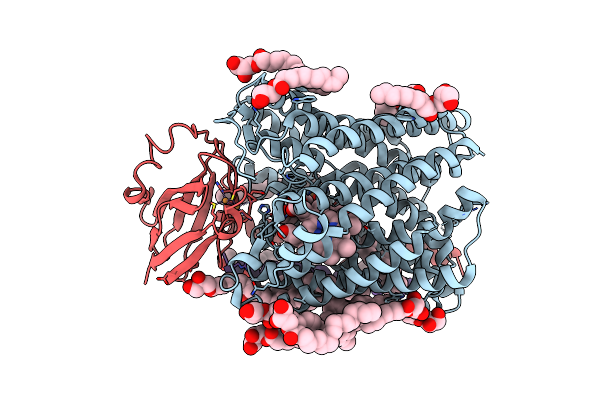
Structure Of Recombinant Cytochrome Ba3 Oxidase Mutant Y133W From Thermus Thermophilus
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2013-01-16 Classification: OXIDOREDUCTASE Ligands: CU, HEM, HAS, PER, OLC, CUA |
 |
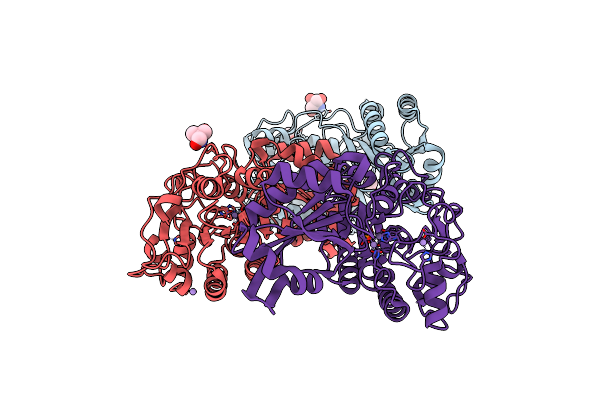
Structure Of Recombinant Cytochrome Ba3 Oxidase Mutant Y133W+T231F From Thermus Thermophilus
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-01-16 Classification: OXIDOREDUCTASE Ligands: CU, HEM, HAS, PER, OLC, CUA |
 |
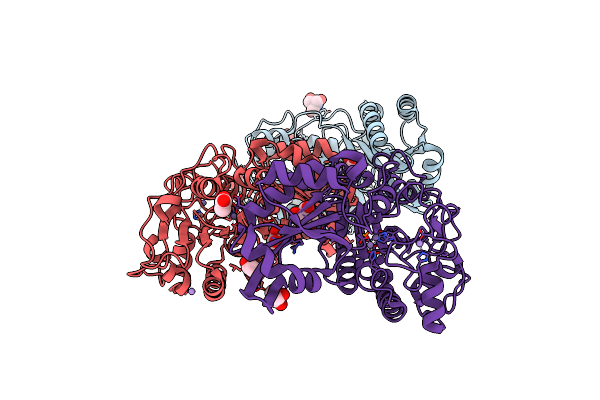
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2010-12-29 Classification: ISOMERASE Ligands: MN, TRS, ACT |
 |
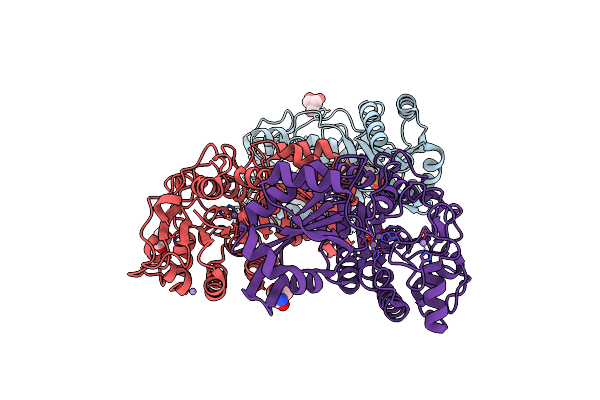
Phosphopentomutase From Bacillus Cereus After Glucose-1,6-Bisphosphate Activation
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-12-29 Classification: ISOMERASE Ligands: MN, ACT, GOL, TRS |
 |
Organism: Bacillus cereus (strain atcc 14579 / dsm 31 / jcm 2152 / nbrc 15305 / ncimb 9373 / nrrl b-3711)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-12-29 Classification: ISOMERASE Ligands: MN, TRS, ACT, GOL, HSX |
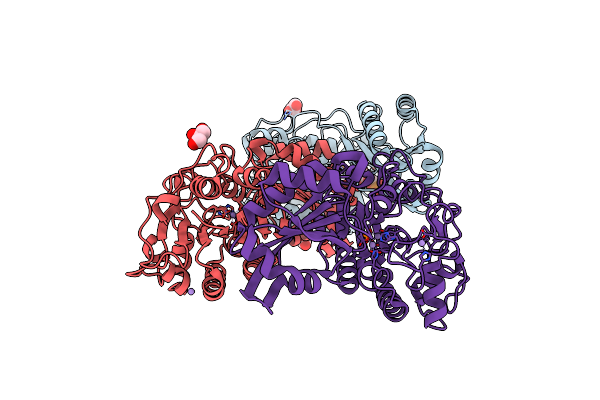 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2010-12-29 Classification: ISOMERASE Ligands: MN, GOL, G16 |
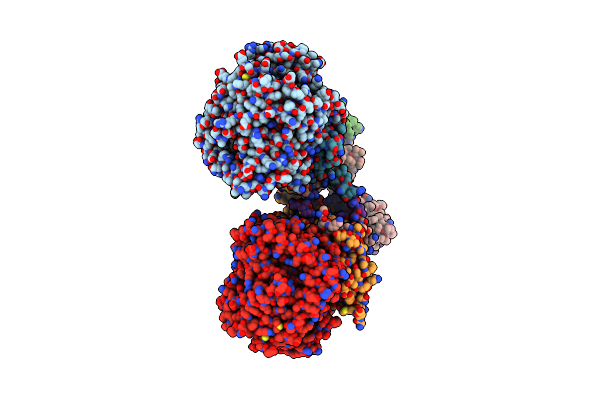 |
Crystal Structure Of Menaquinol:Oxidoreductase In Complex With Oxaloacetate
Organism: Escherichia coli 042, Escherichia coli 536, Escherichia coli dh1
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2010-12-08 Classification: OXIDOREDUCTASE Ligands: FAD, OAA, FES, F3S, SF4 |
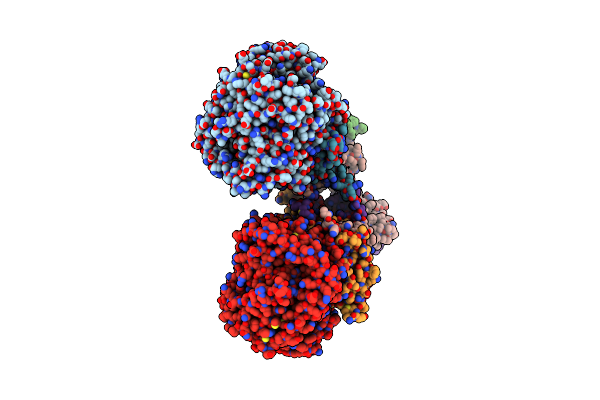 |
Crystal Structure Of Menaquinol:Fumarate Oxidoreductase In Complex With A 3-Nitropropionate Adduct
Organism: Escherichia coli 042, Escherichia coli o55:h7 str. cb9615
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2010-12-08 Classification: OXIDOREDUCTASE Ligands: FAD, 3NP, FES, F3S, SF4 |
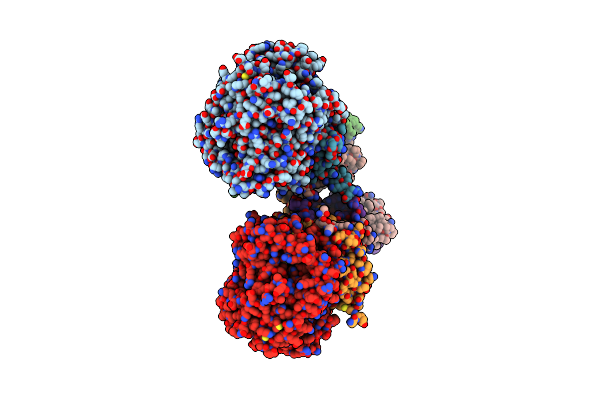 |
Crystal Structure Of Menaquinol:Fumarate Oxidoreductase In Complex With Fumarate
Organism: Escherichia coli 042, Escherichia coli 536, Escherichia coli dh1
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-12-01 Classification: OXIDOREDUCTASE Ligands: FAD, FUM, FES, F3S, SF4 |
 |
Crystal Structure Of Menaquinol:Fumarate Oxidoreductase In Complex With Glutarate
Organism: Escherichia coli 042, Escherichia coli o55:h7
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2010-11-24 Classification: OXIDOREDUCTASE Ligands: FAD, GUA, FES, F3S, SF4 |

