Search Count: 14
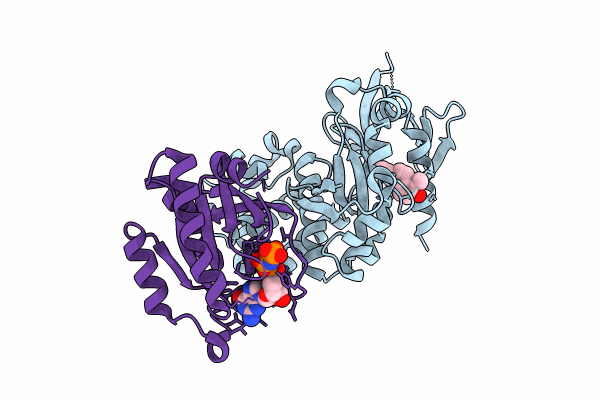 |
Co-Crystal Structure Of Krit1 With A 1-Hydroxy 2-Naphthaldehyde Derivative (6-Ethynyl-2-Hydroxy-1-Naphthaldehyde)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-12-13 Classification: CELL ADHESION Ligands: XHZ, MG, GNP |
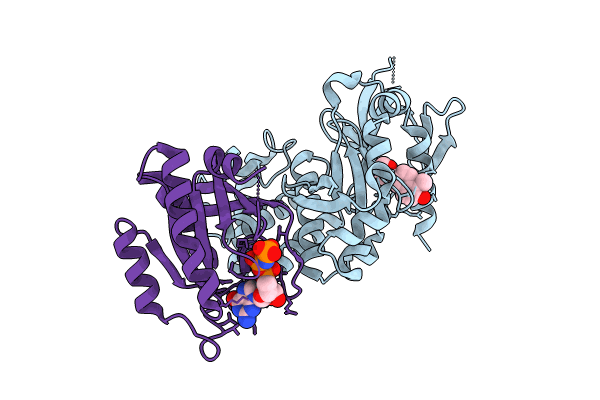 |
Co-Crystal Structure Of Krit1 With A 1-Hydroxy 2-Naphthaldehyde Derivative (6-(Furan-2-Yl)-2-Hydroxy-1-Naphthaldehyde).
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2023-12-06 Classification: CELL ADHESION Ligands: XE2, MG, GNP |
 |
Co-Crystal Structure Of Krit1 With A 1-Hydroxy 2-Naphthaldehyde Derivative (6-(Furan-2-Yl)-2-Hydroxy-1-Naphthaldehyde)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-12-06 Classification: CELL ADHESION Ligands: ZTA, MG, GNP |
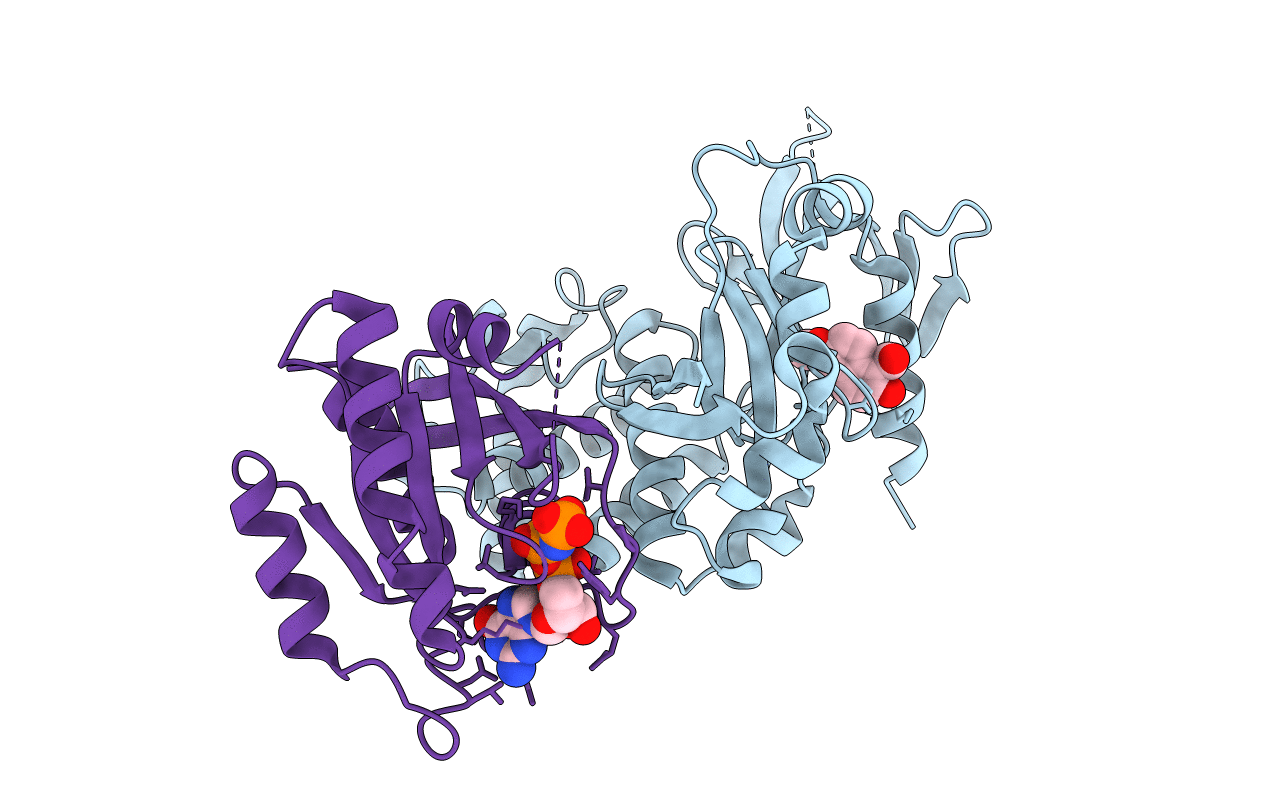 |
Crystal Structure Of The Ternary Complex Of Krit1 Bound To Both The Rap1 Gtpase And Hki6
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2020-11-18 Classification: CELL ADHESION Ligands: QMA, MG, GNP |
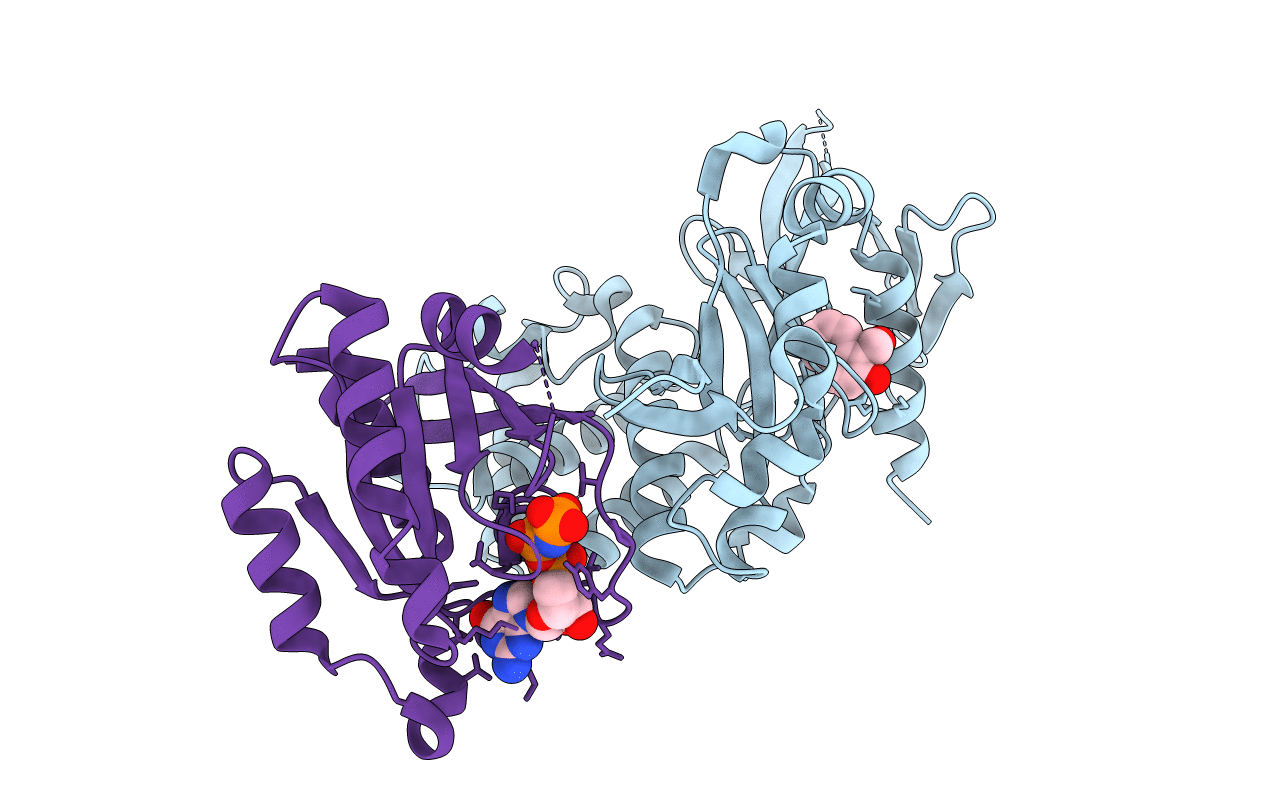 |
Crystal Structure Of The Ternary Complex Of Krit1 Bound To Both The Rap1 Gtpase And Hki2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-06-24 Classification: CELL ADHESION Ligands: 7WO, MG, GNP |
 |
Crystal Structure Of The Ternary Complex Of Krit1 Bound To Both The Rap1 Gtpase And Hki1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-06-24 Classification: CELL ADHESION Ligands: N0G, MG, GNP |
 |
Crystal Structure Of Pbp3 In Complex With Compound 14 ((2E)-2-({[(2S)-2-{[(2Z)-2-(2-Amino-1,3-Thiazol-4-Yl)-2-{[(1,5-Dihydroxy-4-Oxo-1,4-Dihydropyridin-2-Yl)Methoxy]Imino}Acetyl]Amino}-3-Oxopropyl]Oxy}Imino)Pentanedioic Acid)
Organism: Pseudomonas aeruginosa pa1r
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-05-07 Classification: transferase/transferase inhibitor Ligands: 2U2 |
 |
Crystal Structure Of Pbp3 In Complex With Bal30072 ((2Z)-2-(2-Amino-1,3-Thiazol-4-Yl)-2-{[(1,5-Dihydroxy-4-Oxo-1,4-Dihydropyridin-2-Yl)Methoxy]Imino}-N-{(2S)-1-Hydroxy-3-Methyl-3-[(Sulfooxy)Amino]Butan-2-Yl}Ethanamide)
Organism: Pseudomonas aeruginosa pa1r
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-05-07 Classification: transferase/transferase inhibitor Ligands: 2U3 |
 |
Crystal Structure Of Pbp1A In Complex With Compound 17 ((4Z,8S,11E,14S)-5-(2-Amino-1,3-Thiazol-4-Yl)-14-(5,6-Dihydroxy-1,3-Dioxo-1,3-Dihydro-2H-Isoindol-2-Yl)-8-Formyl-2-Methyl-6-Oxo-3,10-Dioxa-4,7,11-Triazatetradeca-4,11-Diene-2,12,14-Tricarboxylic Acid)
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2014-05-07 Classification: transferase/transferase inhibitor Ligands: 2U4 |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-08-21 Classification: Penicillin Binding Protein/Antibiotic Ligands: PFV |
 |
Crystal Structure Of Compound 4D Bound To Large Ribosomal Subunit (50S) From Deinococcus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2013-03-06 Classification: RIBOSOME/RIBOSOME INHIBITOR Ligands: MG, 1F2 |
 |
Crystal Structure Of Compound 4E Bound To Large Ribosomal Subunit (50S) From Deinococcus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2013-03-06 Classification: RIBOSOME/RIBOSOME INHIBITOR Ligands: MG, 1F3 |
 |
Crystal Structure Of Compound 4F Bound To Large Ribosomal Subunit (50S) From Deinococcus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2013-03-06 Classification: RIBOSOME/RIBOSOME INHIBITOR Ligands: MG, 1F4 |
 |
Crystal Structure Of Pseudomonas Aeruginosa Pbp3 Complexed With Compound 14
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-10-17 Classification: PENICILLIN-BINDING PROTEIN/ANTIBIOTIC Ligands: 0W0 |

