Search Count: 13
 |
Organism: Homo sapiens, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-12-13 Classification: STRUCTURAL PROTEIN Ligands: CA, NA, MG, 1PE, GOL, CL |
 |
Organism: Homo sapiens, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2023-12-13 Classification: STRUCTURAL PROTEIN Ligands: CA, NA, MG, 1PE, GOL, CL |
 |
Organism: Homo sapiens, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-12-13 Classification: STRUCTURAL PROTEIN Ligands: CA, NA, 1PE, GOL, CL |
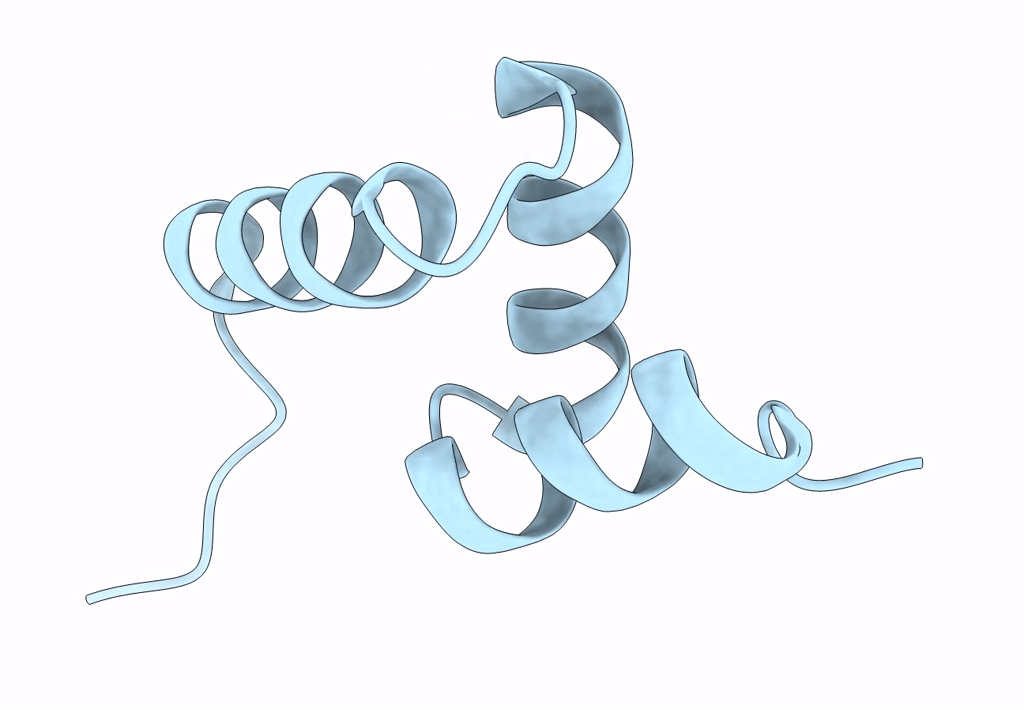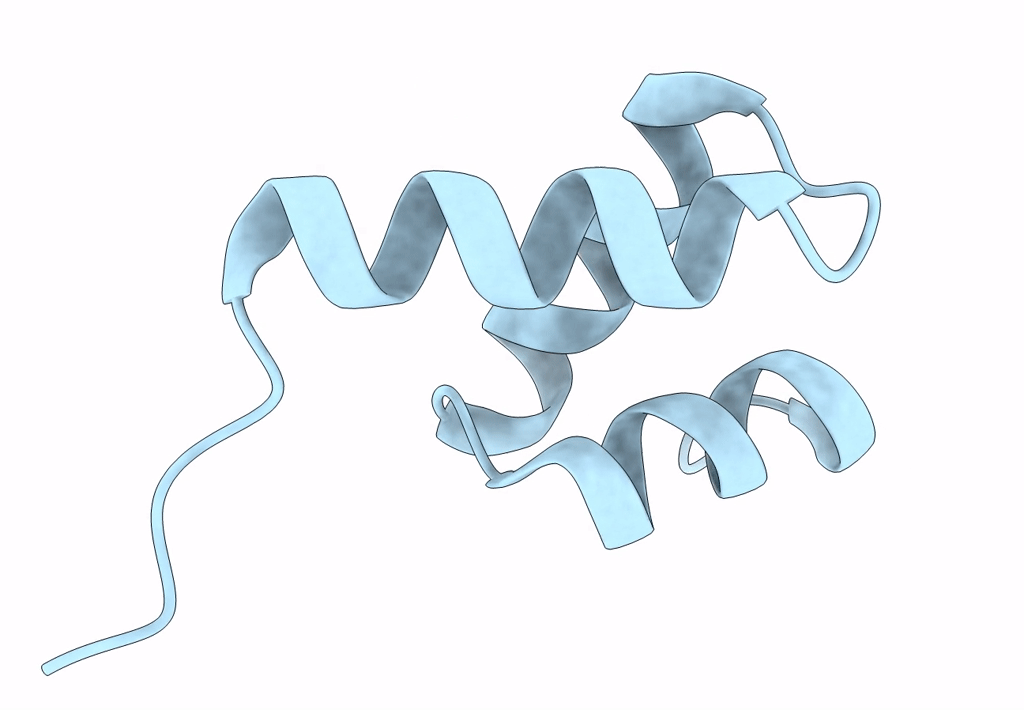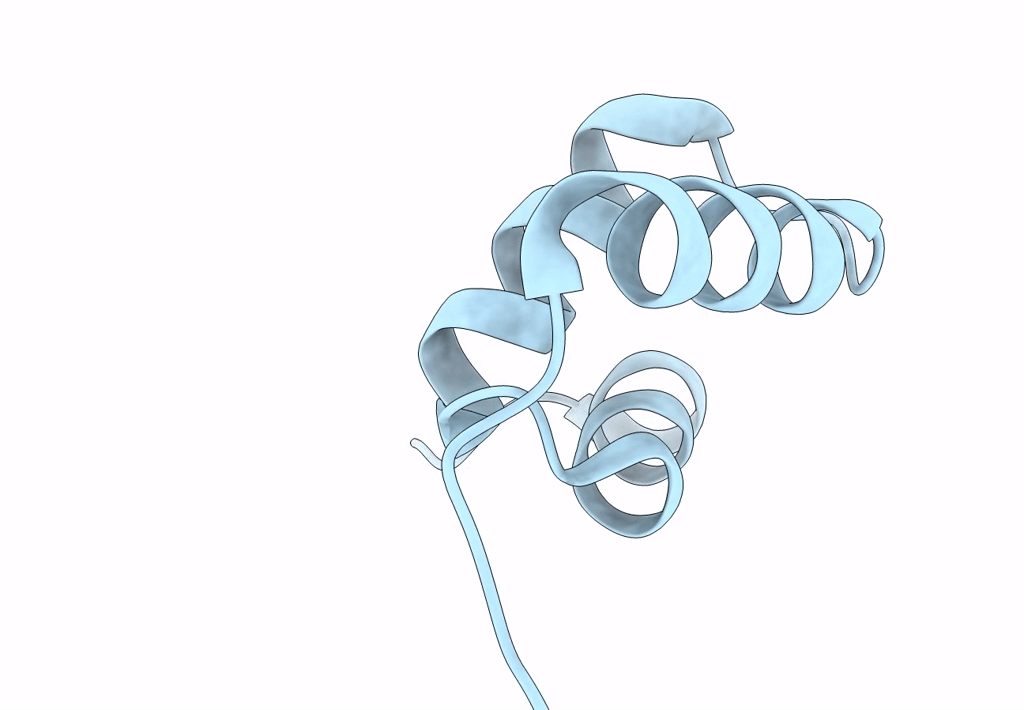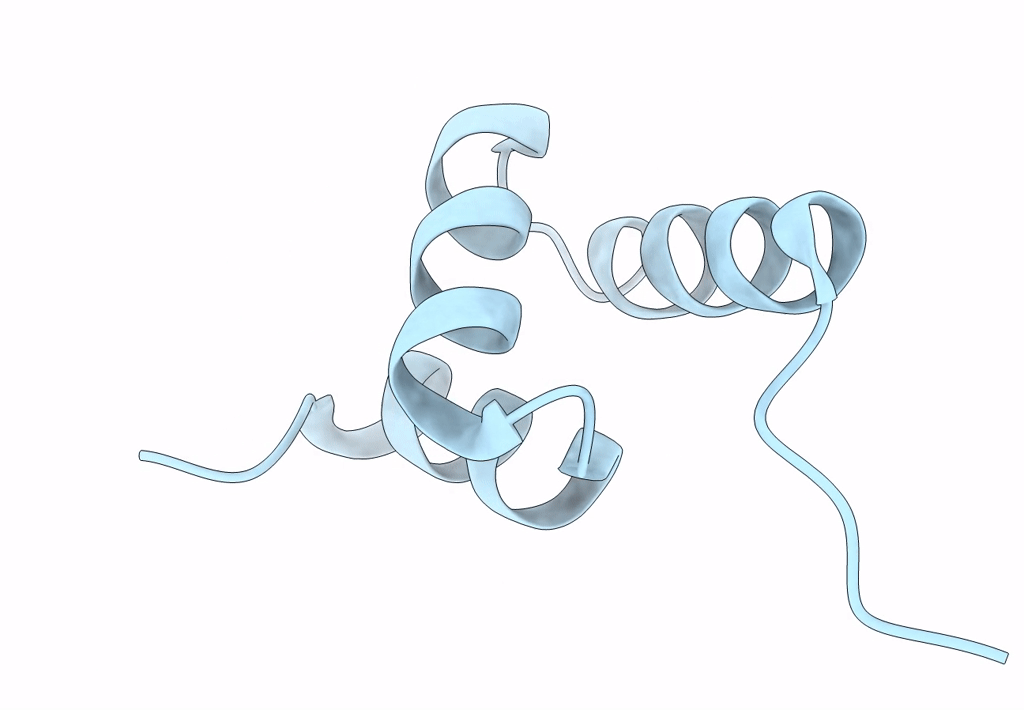 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2023-04-12 Classification: DNA BINDING PROTEIN |
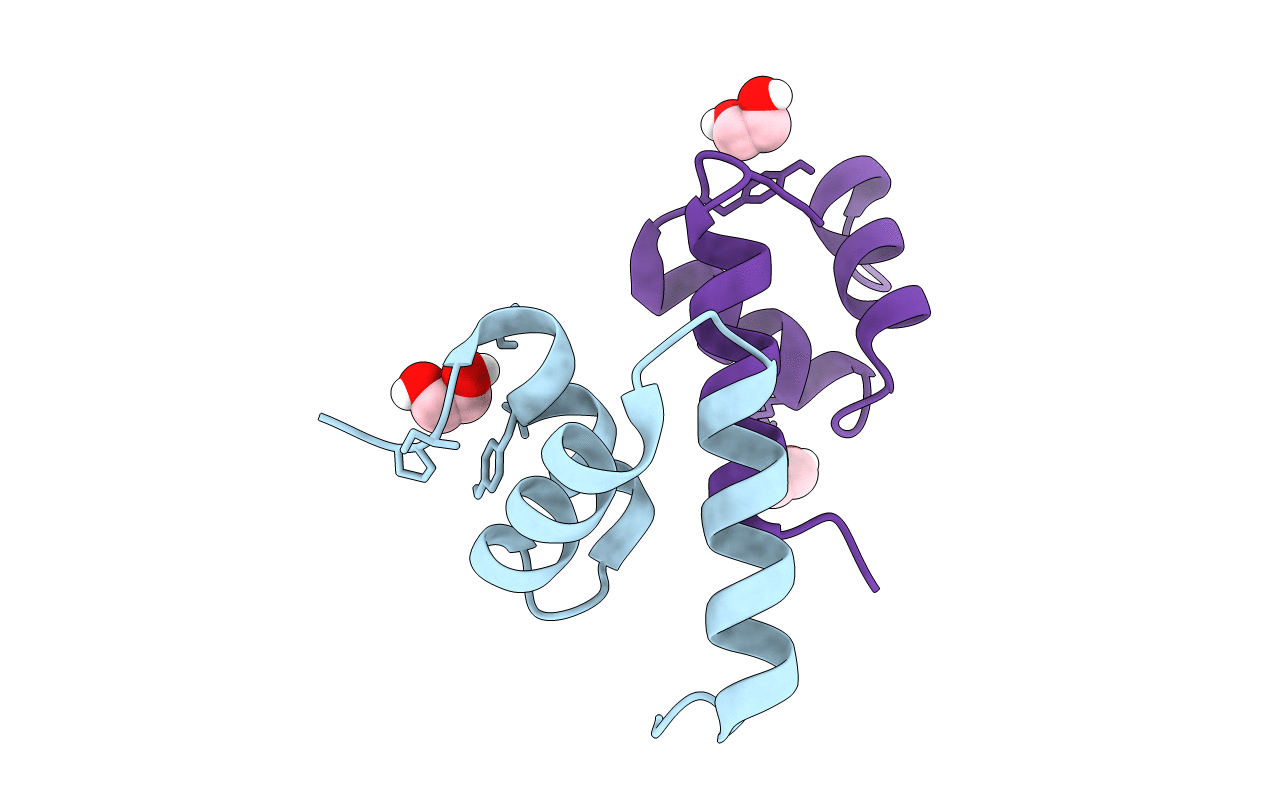 |
Crystal Structure Of Y188G Variant Of The Internal Uba Domain Of Hhr23A In Monoclinic Unit Cell
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2021-03-10 Classification: DNA BINDING PROTEIN Ligands: EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-03-10 Classification: DNA BINDING PROTEIN Ligands: GOL |
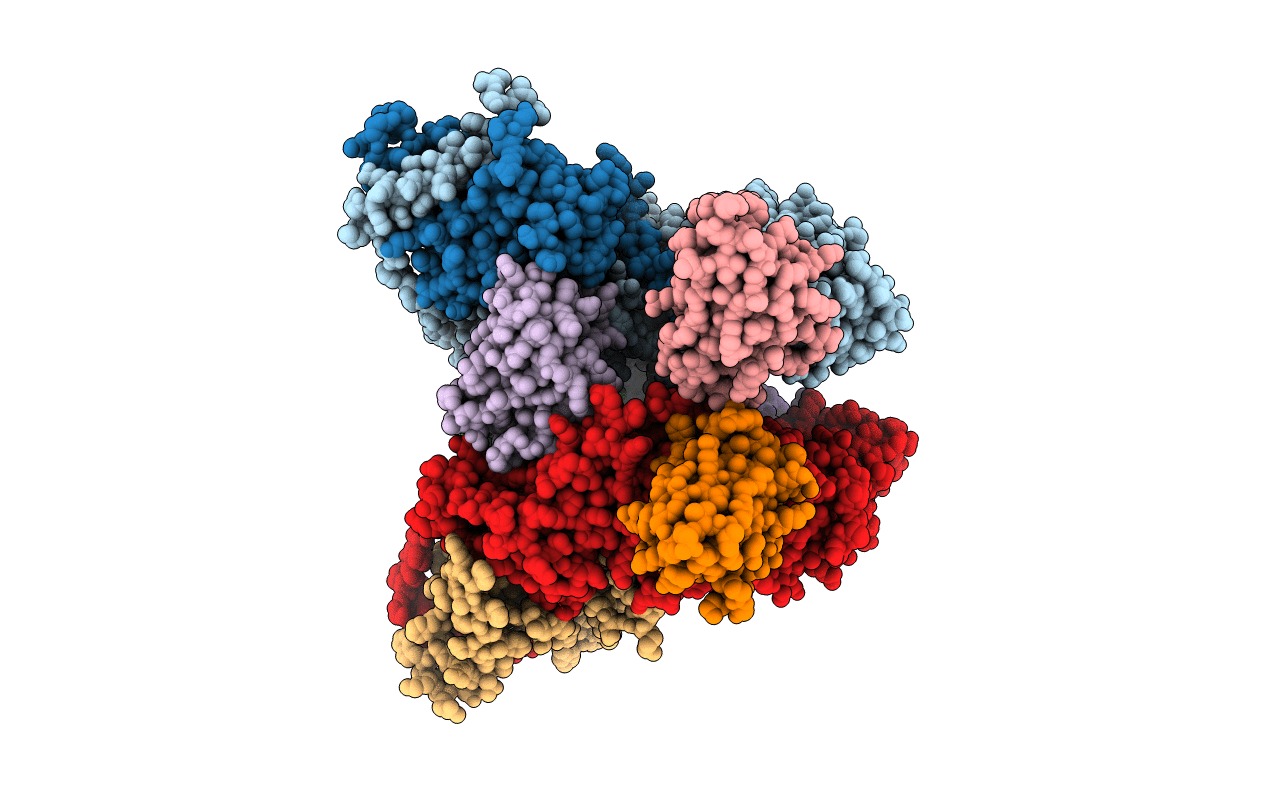 |
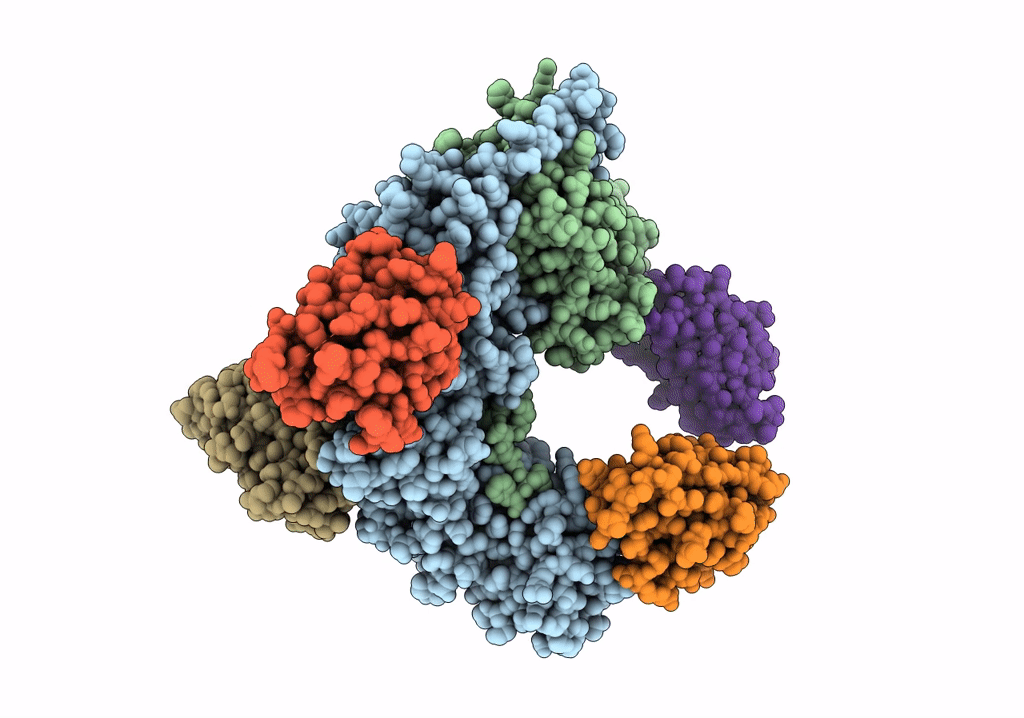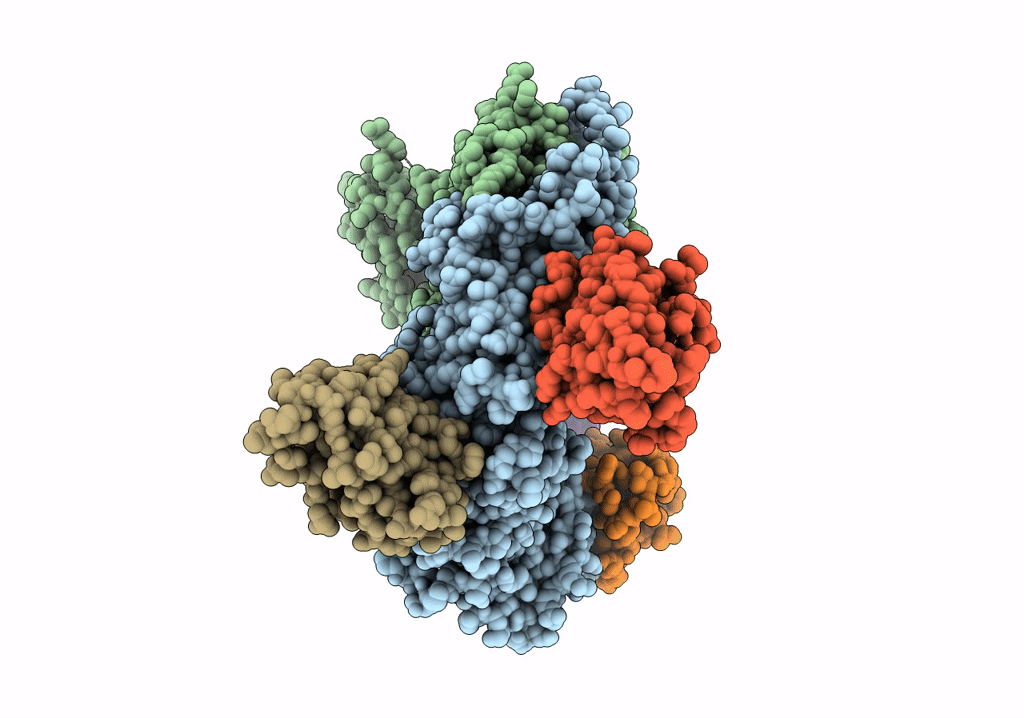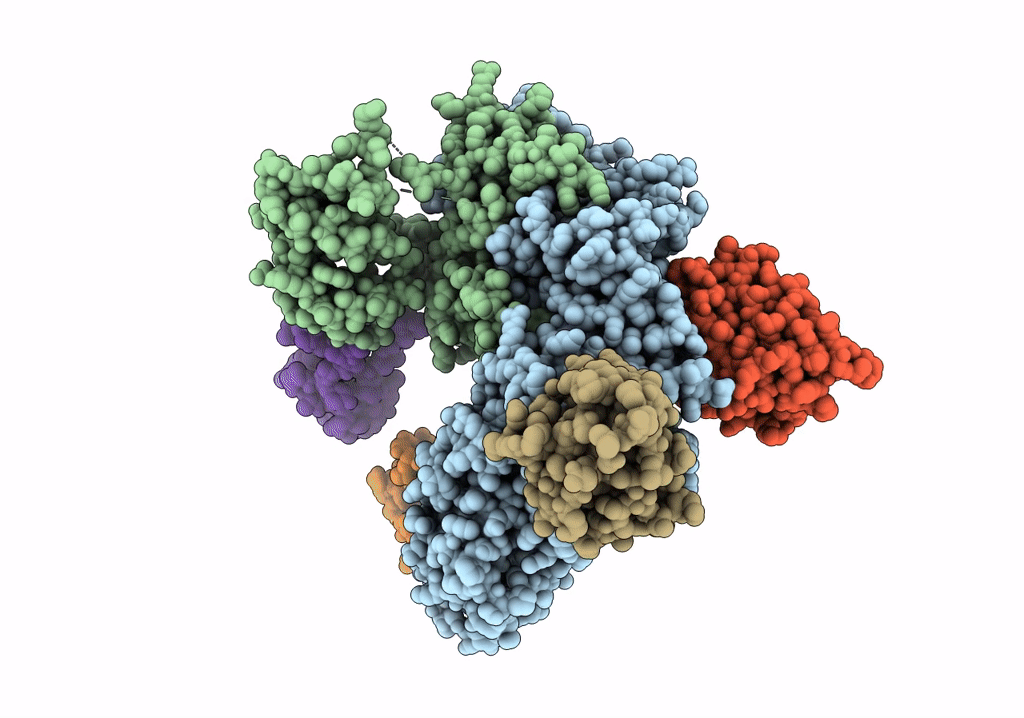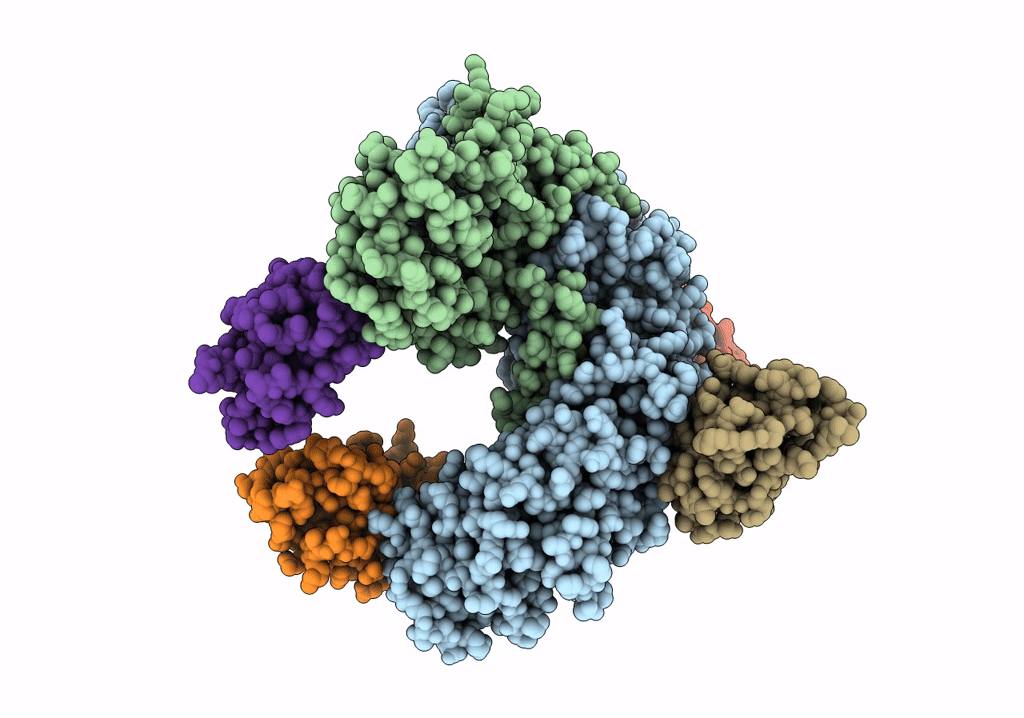
Organism: Rattus norvegicus, Lama glama
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2020-03-11 Classification: SIGNALING PROTEIN |
 |
Organism: Rattus norvegicus, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2020-03-11 Classification: SIGNALING PROTEIN |
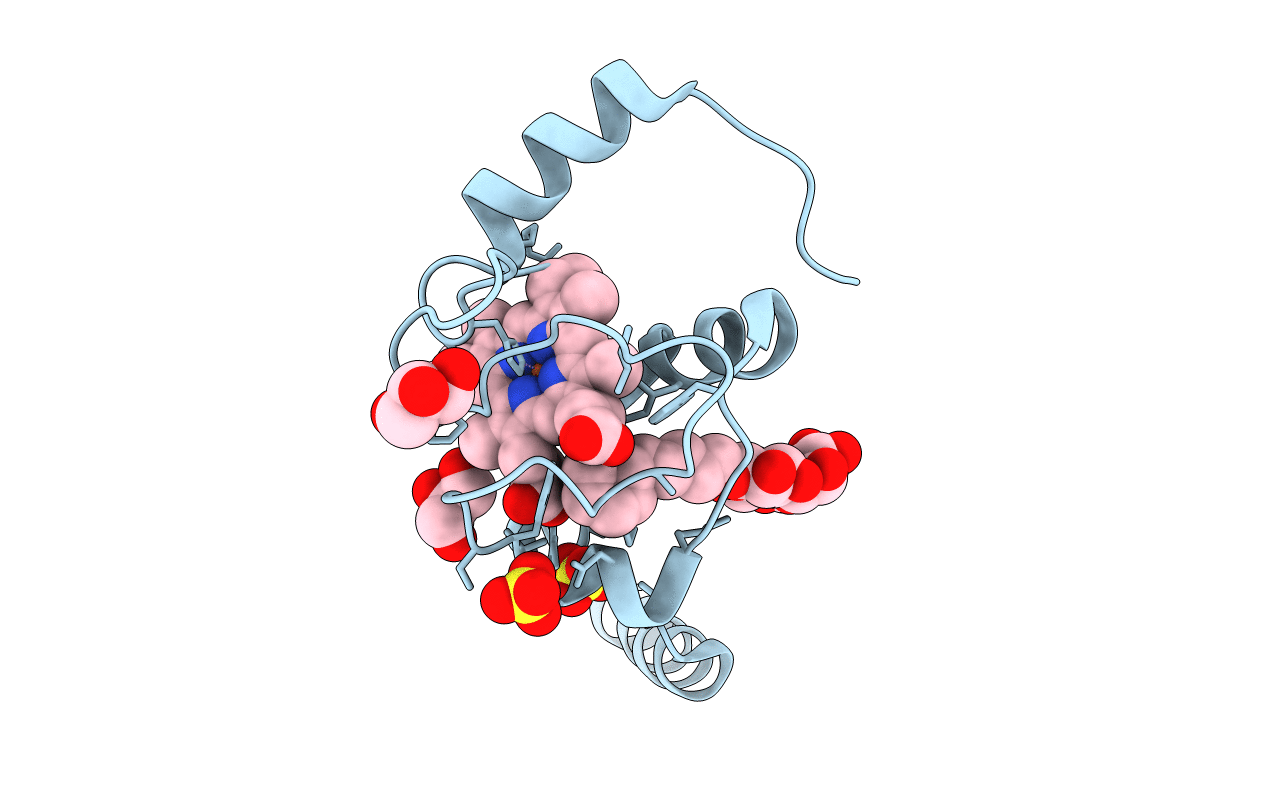 |
Crystal Structure Of A Domain-Swapped Dimer Of Yeast Iso-1-Cytochrome C With Cymal5
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-03-22 Classification: ELECTRON TRANSPORT Ligands: HEC, SO4, CM5, CL, GOL |
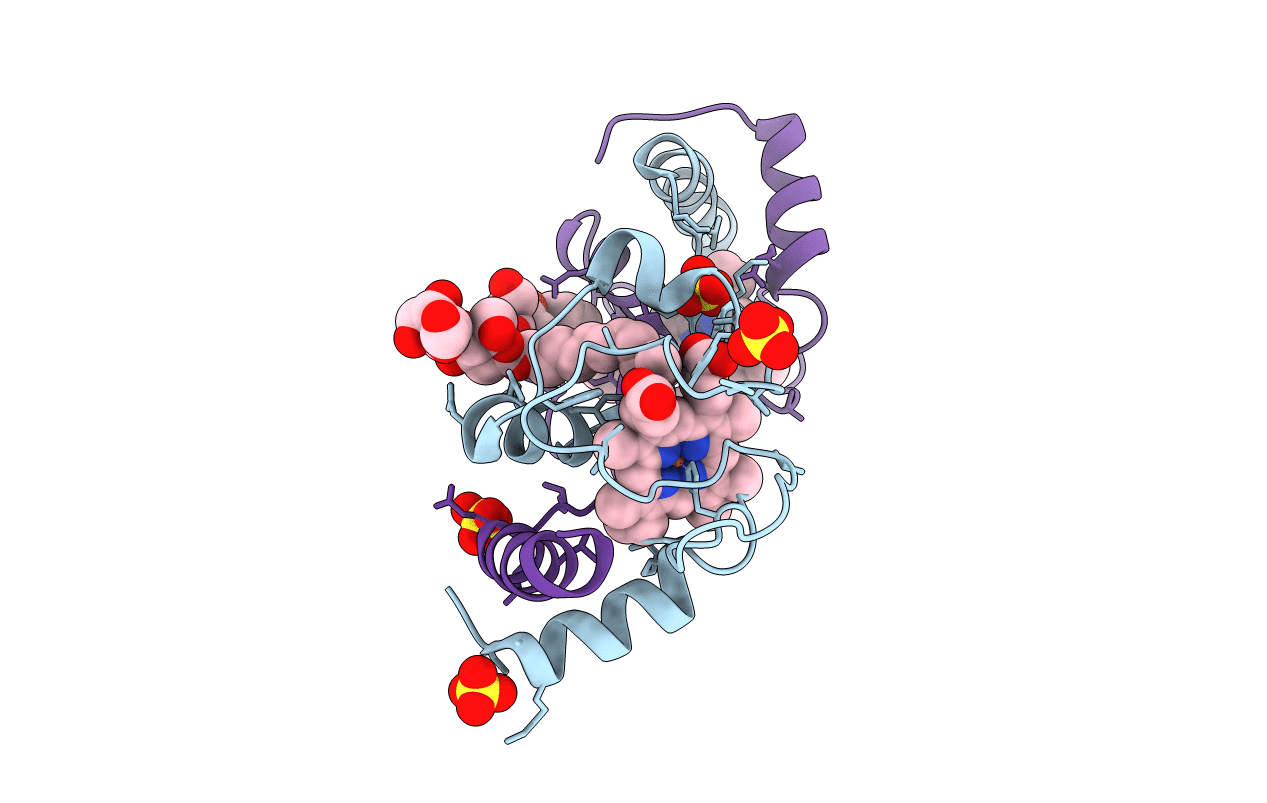 |
Crystal Structure Of A Domain-Swapped Dimer Of Yeast Iso-1-Cytochrome C With Omega-Undecylenyl-Beta-D-Maltopyranoside
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2017-03-22 Classification: ELECTRON TRANSPORT Ligands: HEC, 6UZ, SO4 |
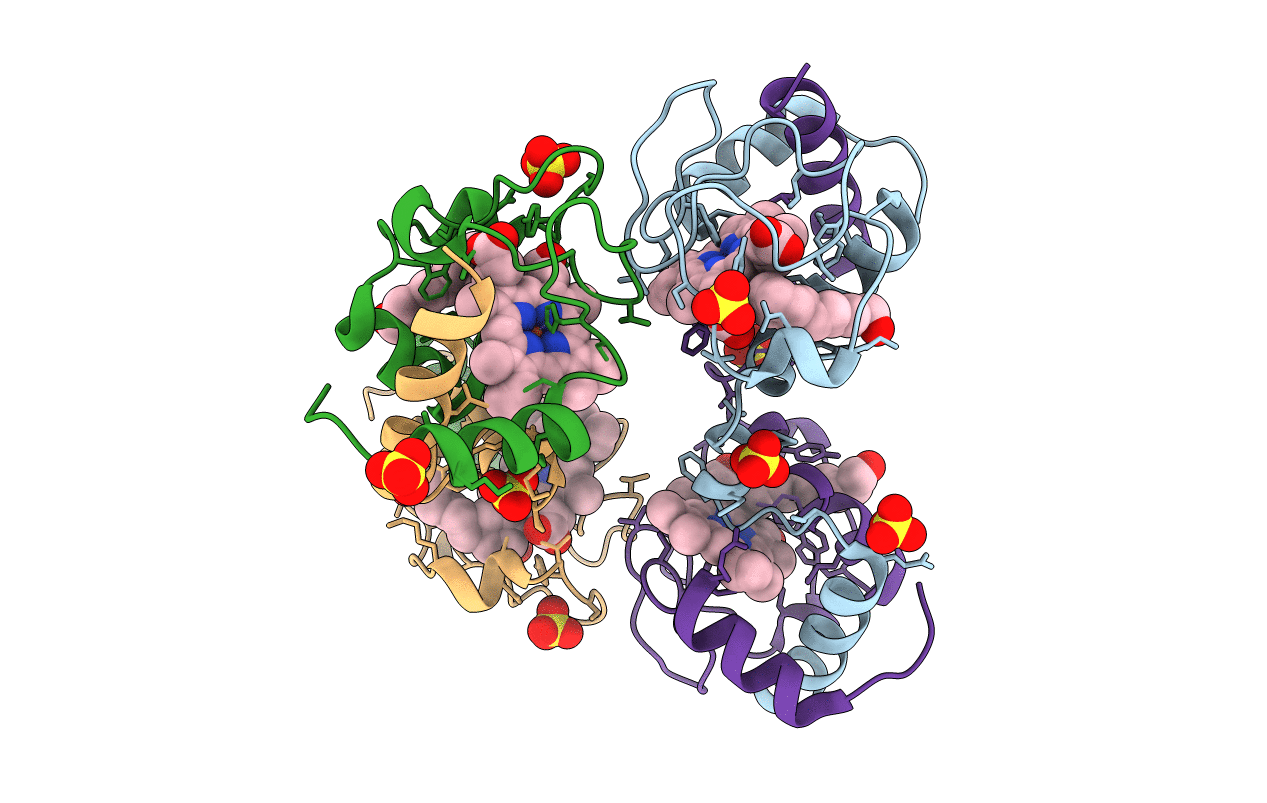 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-03-22 Classification: ELECTRON TRANSPORT Ligands: SO4, ZE7, HEC |
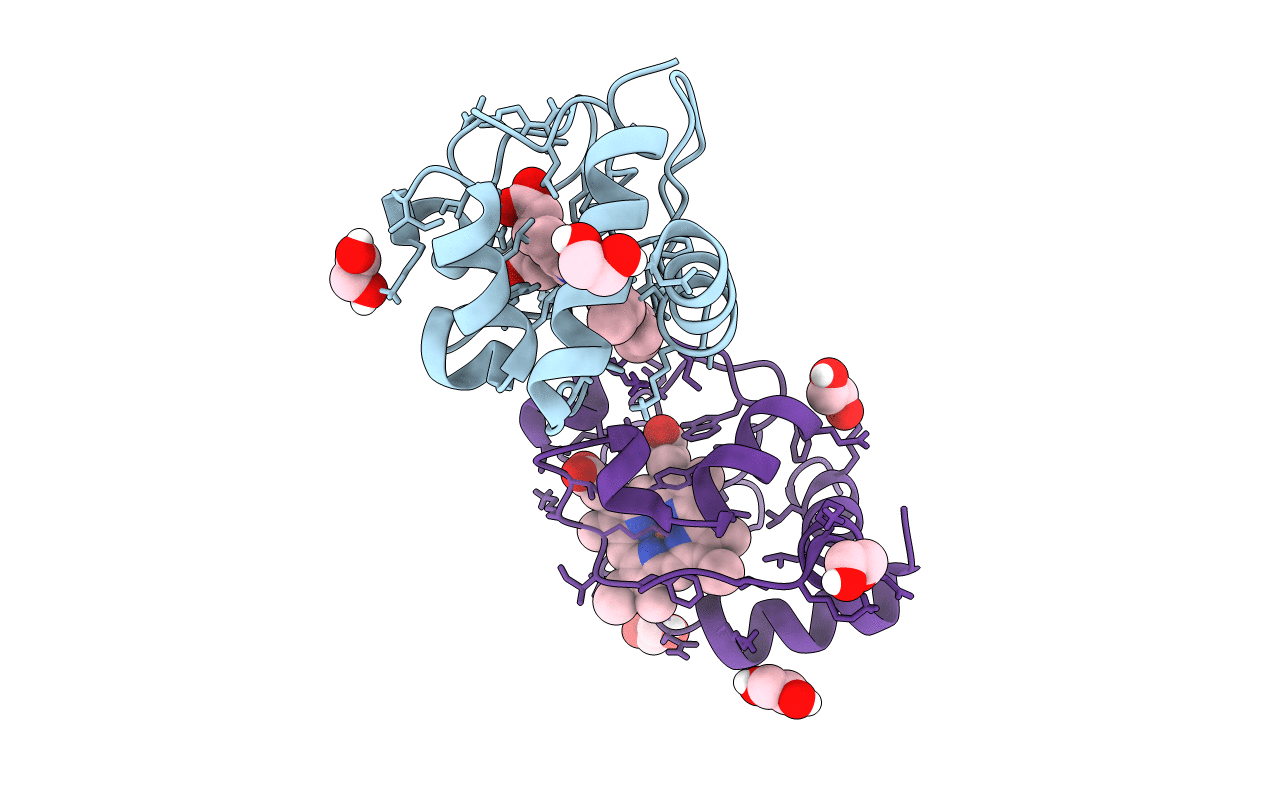 |
Organism: Ateles sp.
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2016-03-02 Classification: ELECTRON TRANSPORT Ligands: HEC, EDO, CL |
 |
Crystal Structure Of An Oxidized Form Of Yeast Iso-1-Cytochrome C At Ph 8.8
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2014-06-04 Classification: ELECTRON TRANSPORT Ligands: HEC, SO4, TBU, GOL |

