Search Count: 27
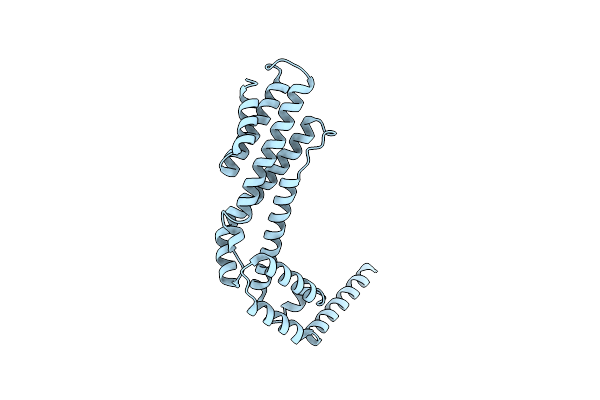 |
Crystal Structure Of The C-Terminal Fragment (Residues 756-982 With The C864S Mutation) Of Arabidopsis Thaliana Chup1
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-01-17 Classification: PLANT PROTEIN |
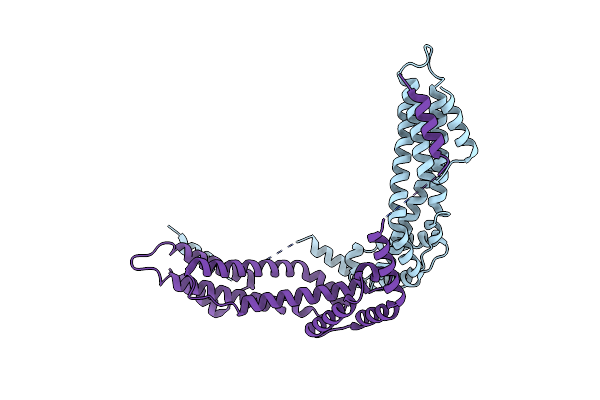 |
Crystal Structure Of The C-Terminal Fragment (Residues 716-982) Of Arabidopsis Thaliana Chup1
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-01-17 Classification: PLANT PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2022-07-20 Classification: TRANSCRIPTION/INHIBITOR Ligands: I0K |
 |
Human Estrogen Receptor Beta Ligand-Binding Domain In Complex With (R)-3-(2-Chloro-4-Hydroxyphenyl)-2-(4-Hydroxyphenyl)Propanenitrile
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2022-07-20 Classification: TRANSCRIPTION/INHIBITOR Ligands: I1D |
 |
Human Estrogen Receptor Beta Ligand-Binding Domain In Complex With (S)-3-(2-Chloro-4-Hydroxyphenyl)-2-(4-Hydroxyphenyl)Propanenitrile
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2022-07-20 Classification: TRANSCRIPTION/INHIBITOR Ligands: I1I |
 |
Human Estrogen Receptor Beta Ligand-Binding Domain In Complex With (R)-2-(2-Chloro-4-Hydroxyphenyl)-3-(4-Hydroxyphenyl)Propanenitrile
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2022-07-20 Classification: TRANSCRIPTION/INHIBITOR Ligands: I1M |
 |
Human Estrogen Receptor Beta Ligand-Binding Domain In Complex With (S)-2-(2-Chloro-4-Hydroxyphenyl)-3-(4-Hydroxyphenyl)Propanenitrile
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2022-07-20 Classification: TRANSCRIPTION/INHIBITOR Ligands: I2B |
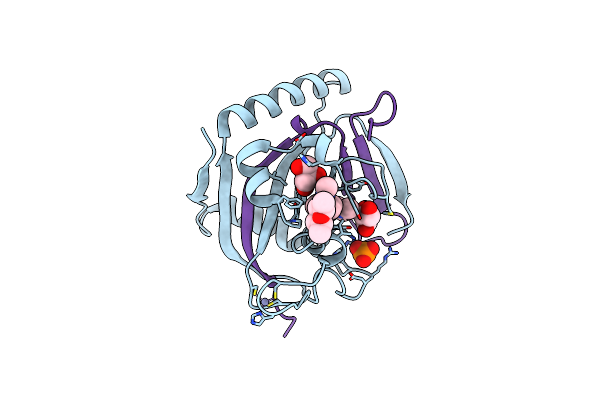 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-04-01 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: DU9, ZN, PO4, GOL |
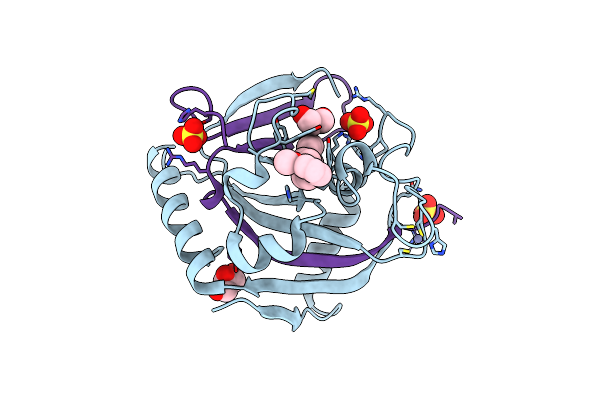 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2019-04-03 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ZN, 9GX, SO4, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2019-04-03 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ZN, 9H3, PO4, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2019-04-03 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ZN, 9H6, PO4, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-04-03 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ZN, 9H9, PO4, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2019-04-03 Classification: TRANSFERASE Ligands: ZN, 9SU, PO4, GOL |
 |
Organism: Halorhodospira halophila
Method: NEUTRON DIFFRACTION Resolution:1.49 Å Release Date: 2017-08-30 Classification: SIGNALING PROTEIN Ligands: HC4, DOD |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-04-30 Classification: CELL ADHESION Ligands: OLC |
 |
Crystal Structure Of The Trans Isomer Of The L93A Mutant Of Bacteriorhodopsin
Organism: Halobacterium
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-06-13 Classification: PROTON TRANSPORT Ligands: RET, SOG, L2P |
 |
Crystal Structure Of The O Intermediate Of The L93A Mutant Of Bacteriorhodopsin
Organism: Halobacterium
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-06-13 Classification: PROTON TRANSPORT Ligands: RET, SOG, L2P |
 |
Organism: Halorhodospira halophila
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2009-03-24 Classification: SIGNALING PROTEIN Ligands: HC4 |
 |
Neutron Crystal Structure Of Photoactive Yellow Protein, Wild Type, At 295K
Organism: Halorhodospira halophila
Method: NEUTRON DIFFRACTION Resolution:1.50 Å Release Date: 2009-03-24 Classification: SIGNALING PROTEIN Ligands: HC4, DOD |
 |
Organism: Pseudechis porphyriacus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-03-11 Classification: TOXIN Ligands: ZN, NA |

