Search Count: 54
 |
Organism: Heteractis crispa
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2018-05-23 Classification: FLUORESCENT PROTEIN Ligands: SO4, 12P, CL |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2015-07-29 Classification: de novo design/DNA |
 |
Using Molecular Dynamics Simulations To Predict Domain Swapping Of Computationally Designed Protein Variants
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2015-05-27 Classification: DE NOVO PROTEIN Ligands: K |
 |
Computational Design And Experimental Verification Of A Symmetric Protein Homodimer
Organism: Drosophila melanogaster
Method: SOLUTION NMR Release Date: 2015-04-08 Classification: DE NOVO PROTEIN |
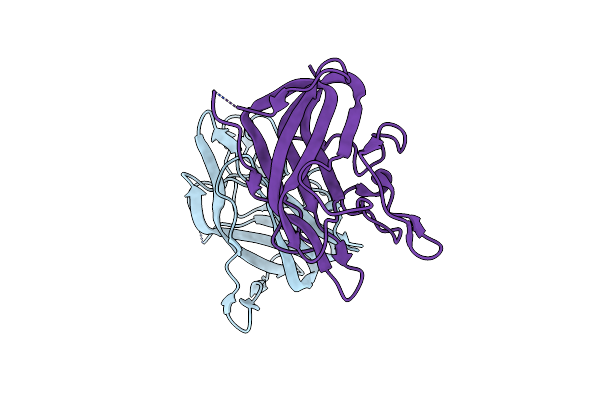 |
The Discobody: An Engineered Discoidin Domain From Factor Viii That Binds V 3 Integrin With Antibody-Like Affinities
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-04-08 Classification: CELL ADHESION |
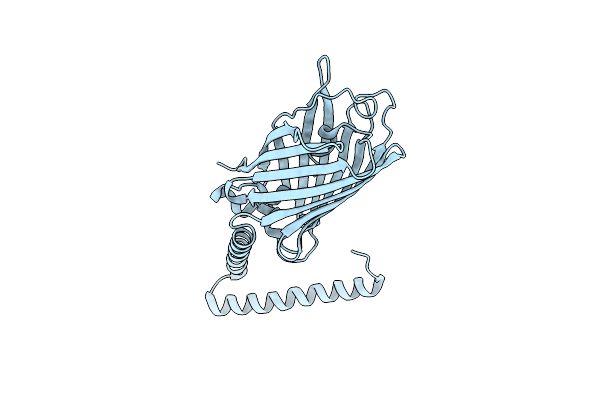 |
Crystal Structure Of A Computational Designed Engrailed Homeodomain Variant Fused With Yfp
Organism: Aequorea victoria, Eimeria acervulina
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-11-05 Classification: fluorescent protein, de novo protein |
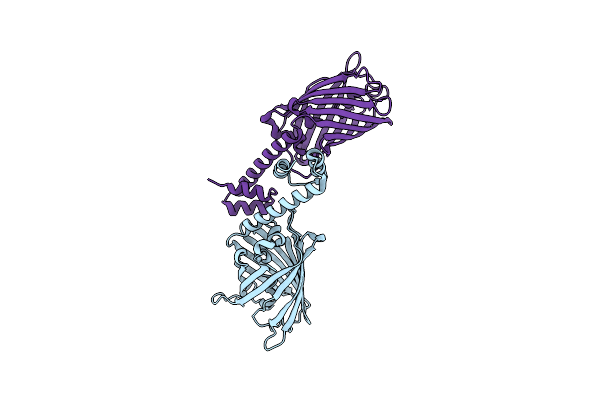 |
Crystal Structure Of A Computational Designed Engrailed Homeodomain Variant Fused With Yfp
Organism: Aequorea victoria, Eimeria acervulina
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-11-05 Classification: fluorescent protein, de novo protein |
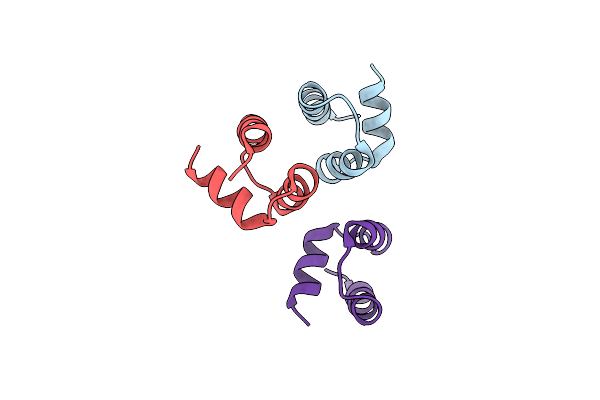 |
Computational Design And Experimental Verification Of A Symmetric Homodimer
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2014-11-05 Classification: DE NOVO PROTEIN Ligands: HOH |
 |
Organism: Actinia equina
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2014-02-26 Classification: FLUORESCENT PROTEIN Ligands: CL |
 |
Crystal Structure Of Kemp Eliminase Hg3.17 E47N,N300D Complexed With Transition State Analog 6-Nitrobenzotriazole
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2013-10-16 Classification: LYASE/LYASE INHIBITOR Ligands: SO4, 6NT |
 |
Organism: Discosoma sp. lw-2004
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2012-10-31 Classification: FLUORESCENT PROTEIN |
 |
Organism: Discosoma sp. lw-2004
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-10-24 Classification: FLUORESCENT PROTEIN |
 |
Organism: Discosoma sp. lw-2004
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2012-10-24 Classification: FLUORESCENT PROTEIN Ligands: CL |
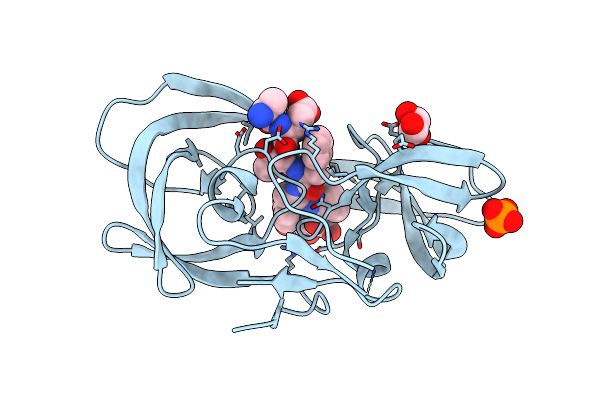 |
Crystal Structure Of Inactive Single Chain Wild-Type Hiv-1 Protease In Complex With The Substrate Rt-Rh
Organism: Hiv-1 m:b_arv2/sf2, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-06-06 Classification: hydrolase/hydrolase substrate Ligands: GOL, PO4 |
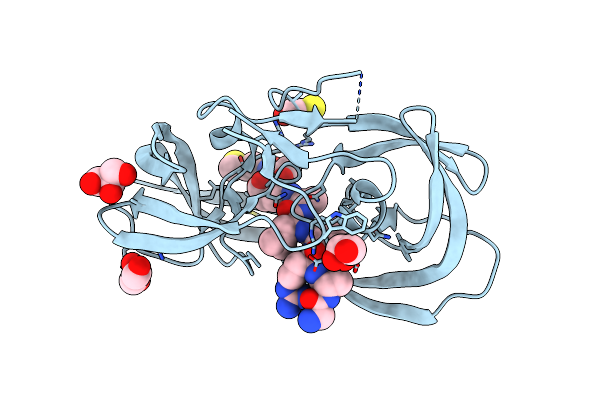 |
Crystal Structure Of Inactive Single Chain Wild-Type Hiv-1 Protease In Complex With The Substrate Ca-P2
Organism: Hiv-1 m:b_arv2/sf2, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2012-06-06 Classification: hydrolase/hydrolase substrate Ligands: GOL, BME |
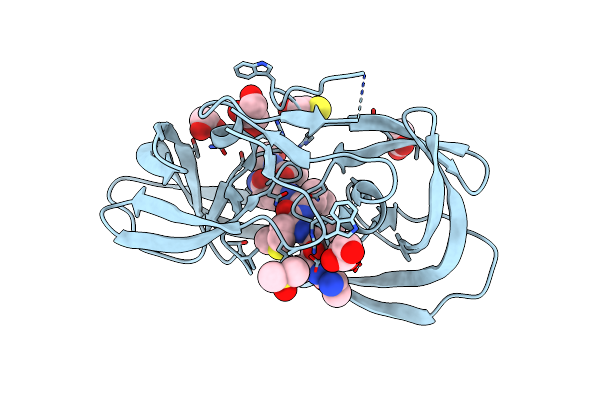 |
Crystal Structure Of Inactive Single Chain Wild-Type Hiv-1 Protease In Complex With The Substrate P2-Nc
Organism: Hiv-1 m:b_arv2/sf2, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2012-06-06 Classification: hydrolase/hydrolase substrate Ligands: GOL, EDO, DMS, ACT, BME |
 |
Crystal Structure Of Inactive Single Chain Variant Of Hiv-1 Protease In Complex With The Substrate P2-Nc
Organism: Hiv-1 m:b_arv2/sf2, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-06-06 Classification: hydrolase/hydrolase substrate Ligands: BME, EDO, GOL, ACT, PO4 |
 |
Crystal Structure Of Inactive Single Chain Variant Of Hiv-1 Protease In Complex With The Substrate Rt-Rh
Organism: Hiv-1 m:b_arv2/sf2, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-06-06 Classification: hydrolase/hydrolase substrate Ligands: EDO, GOL |
 |
Crystal Structure Of Cel5A (Eg2) From Hypocrea Jecorina (Trichoderma Reesei)
Organism: Hypocrea jecorina
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2011-11-02 Classification: HYDROLASE Ligands: SO4, MG |
 |
Organism: Nostoc ellipsosporum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-08-03 Classification: ANTIVIRAL PROTEIN |

