Search Count: 88
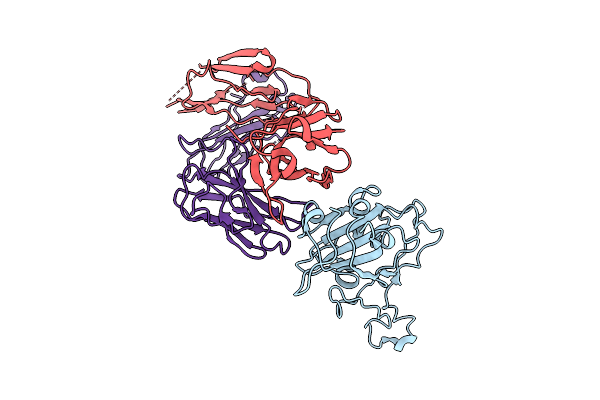 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-171
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
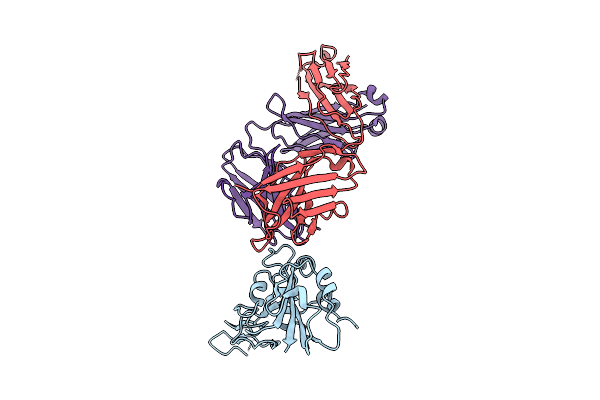
 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-183
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
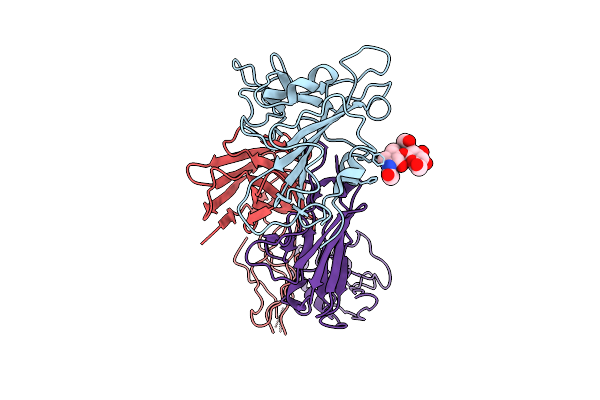
 |
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-171
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
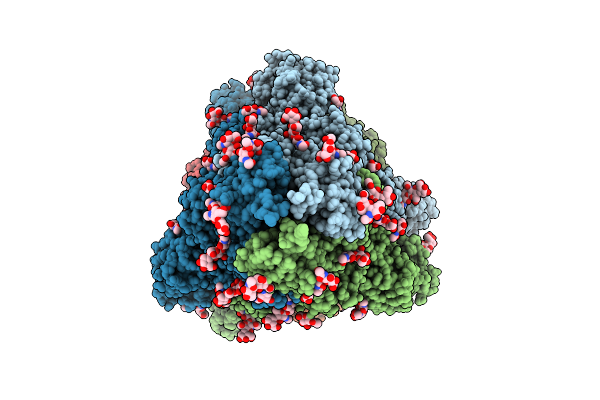
 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-198
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
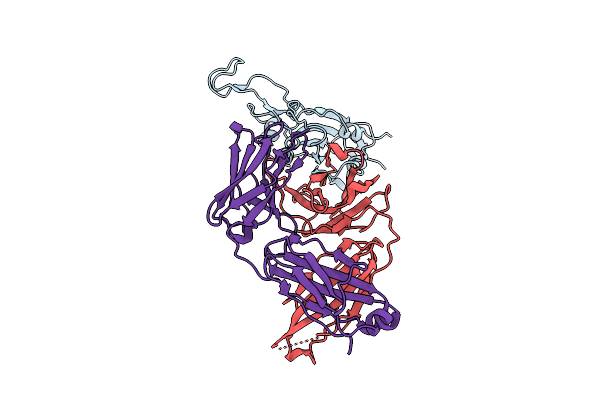 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-203
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
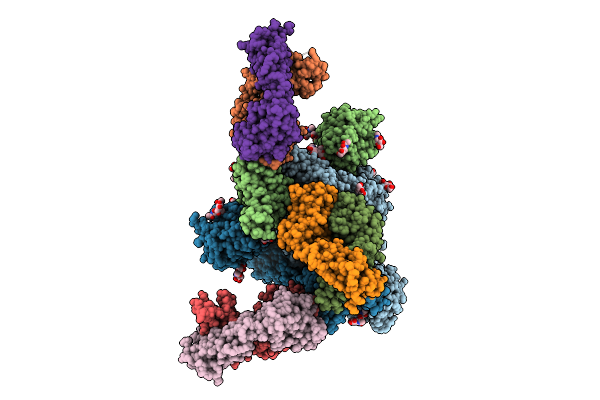 |
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-203
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
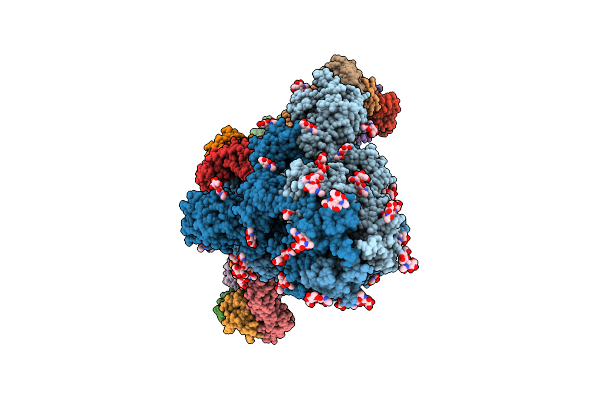 |
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-198
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
N-Acetylglucosamine 6-Phosphate Dehydratase: Inhibited 6-Phosphogluconic Acid State Of Nags
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2025-02-19 Classification: SUGAR BINDING PROTEIN Ligands: SO4, 6PG |
 |
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-02-19 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
 |
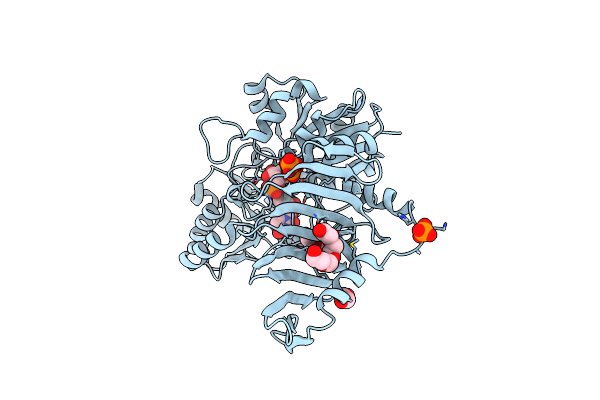
N-Acetylglucosamine 6-Phosphate Dehydratase: Glcnac6P Substrate-Bound State Of Nags
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2025-02-19 Classification: SUGAR BINDING PROTEIN Ligands: SO4, 16G |
 |
Organism: Staphylococcus epidermidis
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2023-12-06 Classification: TRANSFERASE Ligands: CL, PO4, UPG, EDO, PG4, PGE |
 |
Organism: Staphylococcus epidermidis
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2023-12-06 Classification: TRANSFERASE Ligands: BJT, CL, BME, UDP, GOL |
 |
Organism: Tannerella forsythia
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-08-02 Classification: TRANSFERASE Ligands: GOL, EDO |
 |
Organism: Tannerella forsythia
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-08-02 Classification: TRANSFERASE Ligands: SO4, GOL |
 |
Crystal Structure Of Tannerella Forsythia Murnac Kinase Murk With A Phosphate Analogue
Organism: Tannerella forsythia
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2023-08-02 Classification: TRANSFERASE Ligands: ACP, SO4, TAM, MG, EDO, PEG |
 |
Crystal Structure Of Tannerella Forsythia Sugar Kinase K1058 In Complex With N-Acetylmuramic Acid (Murnac)
Organism: Tannerella forsythia
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2023-08-02 Classification: TRANSFERASE Ligands: AMU, SO4 |
 |
Crystal Structure Of Tannerella Forsythia Murnac Kinase Murk In Complex With N-Acetylmuramic Acid (Murnac)
Organism: Tannerella forsythia
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-08-02 Classification: TRANSFERASE Ligands: AMU |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2023-05-10 Classification: TRANSFERASE Ligands: CL, PO4, GOL, 1PE |
 |
Organism: Staphylococcus epidermidis
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2023-05-10 Classification: TRANSFERASE Ligands: CWI, EDO, CL |
 |
Organism: Staphylococcus epidermidis
Method: X-RAY DIFFRACTION Resolution:3.21 Å Release Date: 2023-05-10 Classification: TRANSFERASE Ligands: CL, BME, EDO |

