Search Count: 22
 |
Cryo-Em Structure Of Aquifex Aeolicus Minimal Protein-Only Rnase P (Harp) In Complex With Pre-Trnas
Organism: Aquifex aeolicus vf5
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: HYDROLASE-RNA COMPLEX |
 |
Cryo-Em Structure Of Hydrogenobacter Thermophilus Minimal Protein-Only Rnase P (Harp) In Complex With Pre-Trnas
Organism: Hydrogenobacter thermophilus dsm 653, Aquifex aeolicus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: HYDROLASE/RNA Ligands: MG |
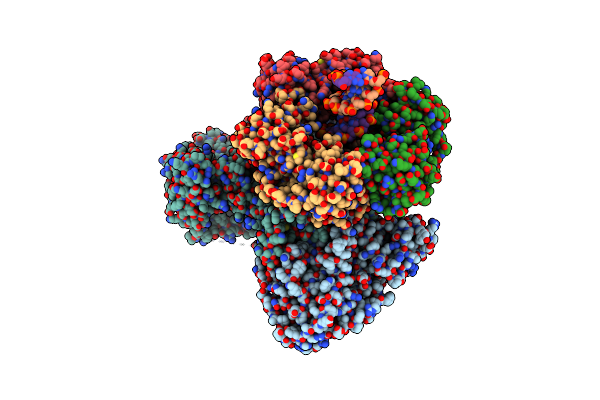 |
Organism: Thermococcus kodakarensis (strain atcc baa-918 / jcm 12380 / kod1), Thermococcus kodakarensis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-08-05 Classification: REPLICATION/DNA Ligands: FE, ZN |
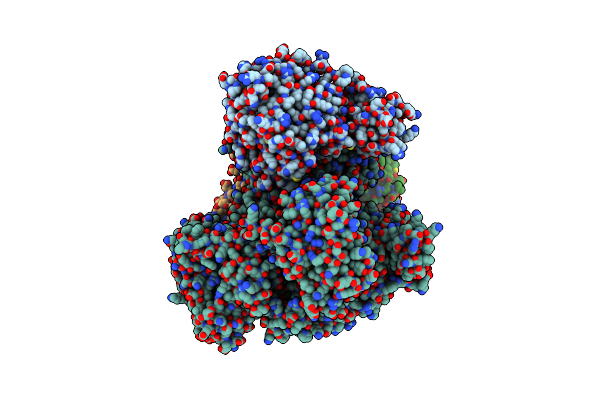 |
Organism: Thermococcus kodakarensis kod1, Thermococcus kodakarensis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-08-05 Classification: REPLICATION/DNA Ligands: FE, ZN |
 |
Organism: Thermococcus kodakarensis kod1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-11-02 Classification: HYDROLASE/DNA Ligands: MG, MPD |
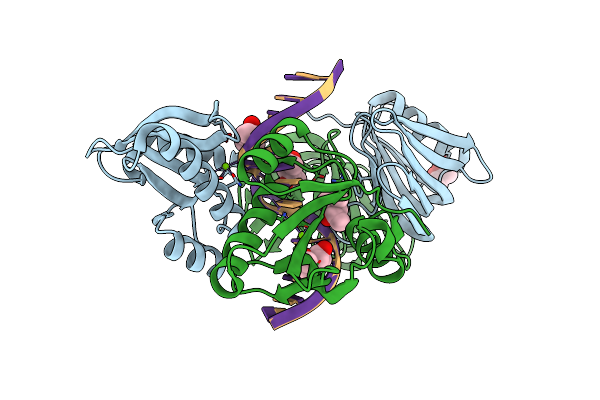 |
Organism: Thermococcus kodakarensis kod1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-11-02 Classification: HYDROLASE/DNA Ligands: MPD, MG |
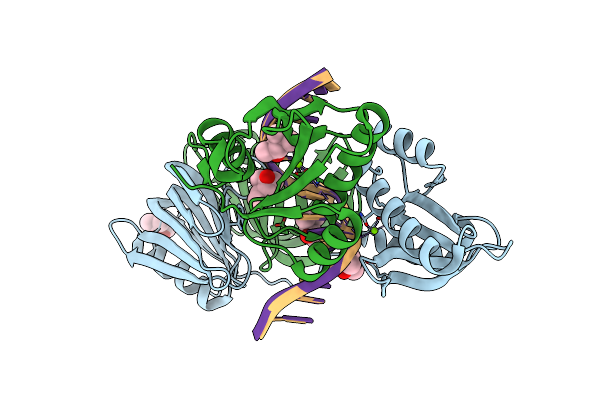 |
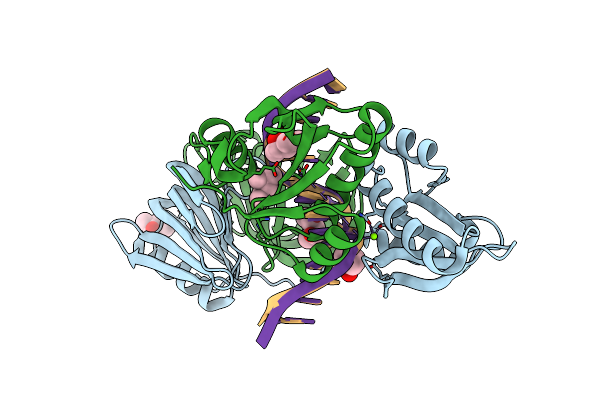
Organism: Thermococcus kodakarensis kod1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-11-02 Classification: HYDROLASE/DNA Ligands: MG, MPD |
 |
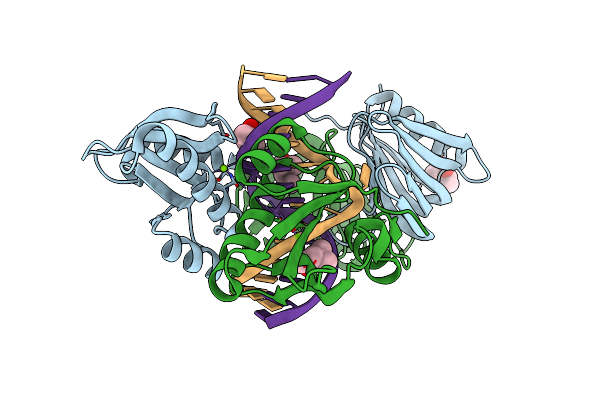
Organism: Thermococcus kodakarensis kod1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2016-11-02 Classification: HYDROLASE/DNA Ligands: MPD, MG |
 |
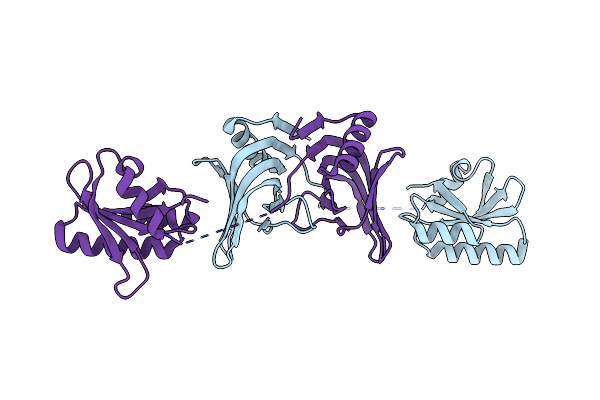
Organism: Thermococcus kodakarensis kod1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2016-11-02 Classification: HYDROLASE/DNA Ligands: MG, MPD |
 |
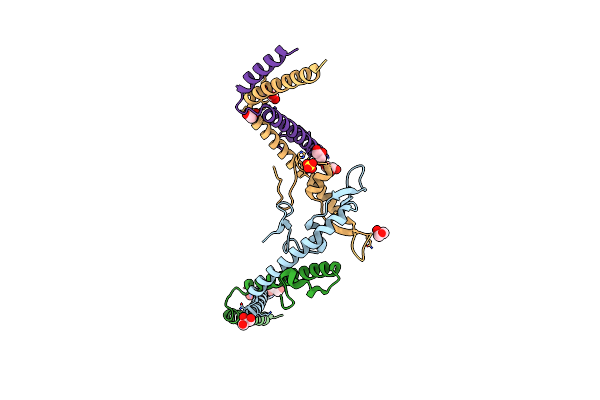
Organism: Thermococcus kodakarensis kod1
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-11-02 Classification: HYDROLASE |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-08-22 Classification: RECOMBINATION ACTIVATOR Ligands: GOL, NO3, SO4 |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-08-22 Classification: RECOMBINATION ACTIVATOR Ligands: BOG |
 |
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2011-07-27 Classification: REPLICATION |
 |
Organism: Vibrio parahaemolyticus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-03-31 Classification: TOXIN |
 |
Crystal Structure Of Pyrococcus Furiosus Dna Polymerase/Pcna Monomer Mutant Complex
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2009-11-03 Classification: TRANSFERASE/REPLICATION |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2009-02-10 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2009-02-10 Classification: HYDROLASE Ligands: SO4, ADP |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-02-10 Classification: HYDROLASE |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2009-02-10 Classification: HYDROLASE Ligands: ACP |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2002-11-06 Classification: HYDROLASE Ligands: ANP |

