Search Count: 38
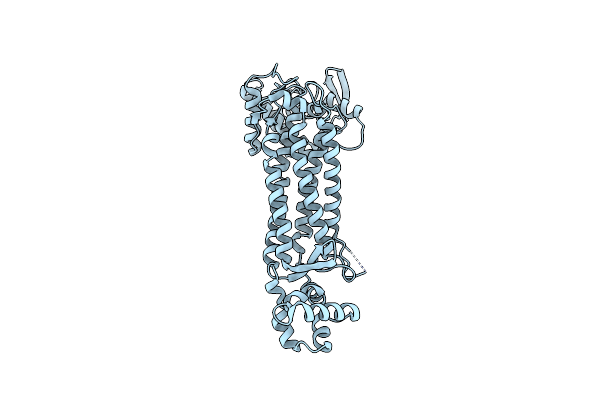 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2023-11-15 Classification: LYASE |
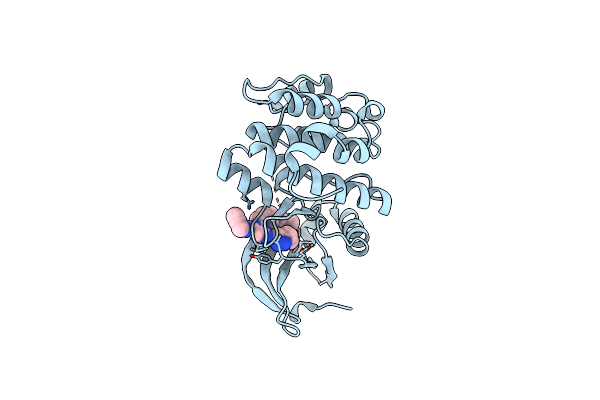 |
Reversible Lysine-Targeted Probes Reveal Residence Time-Based Kinase Selectivity In Vivo
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2022-06-08 Classification: CELL CYCLE Ligands: 5YZ, CL |
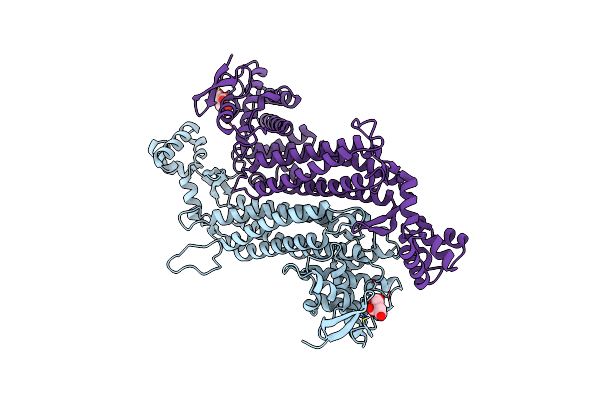 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2020-05-27 Classification: LYASE Ligands: FLC |
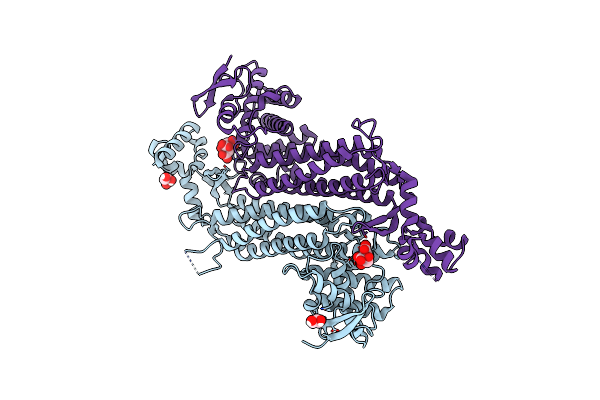 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2020-05-06 Classification: LYASE Ligands: FLC, GOL |
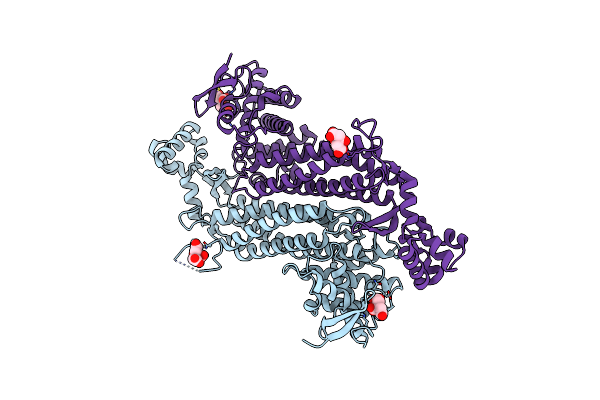 |
Crystal Structure Of E. Coli Fumarase C Bound To Citrate At 1.53 Angstrom Resolution
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2019-09-25 Classification: LYASE Ligands: CIT |
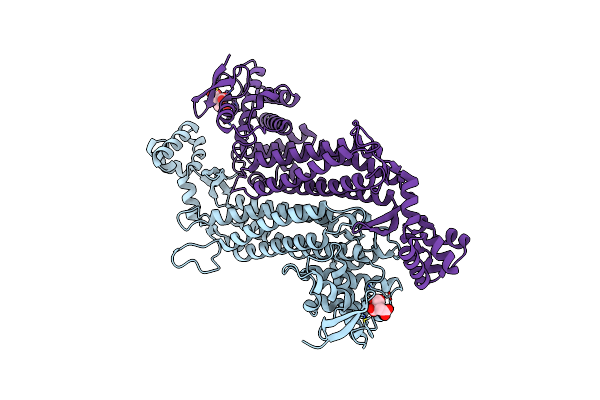 |
Crystal Structure Of E. Coli Fumarase C K324A Variant With Closed Ss Loop At 1.41 Angstrom Resolution
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2019-09-25 Classification: LYASE Ligands: CIT |
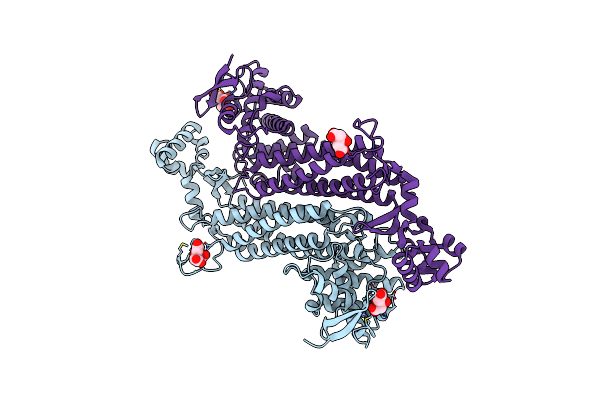 |
Crystal Structure Of E. Coli Fumarase C S318A Variant With Closed Ss Loop At 1.37 Angstrom Resolution
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2019-09-25 Classification: LYASE Ligands: CIT |
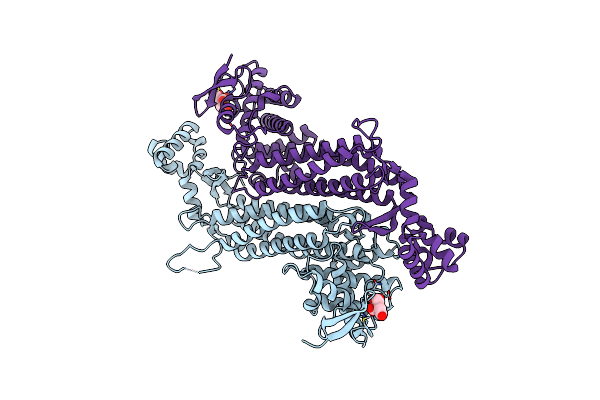 |
Crystal Structure Of E. Coli Fumarase C N326A Variant With Closed Ss Loop At 1.40 Angstrom Resolution
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-09-25 Classification: LYASE Ligands: CIT |
 |
Crystal Structure Of Dcrb From Salmonella Enterica At 1.92 Angstroms Resolution
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2018-10-31 Classification: UNKNOWN FUNCTION |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-12-06 Classification: lipid transport/activator Ligands: ADP, MG, NOV |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-12-06 Classification: lipid transport/activator Ligands: MG, CZJ, ADP |
 |
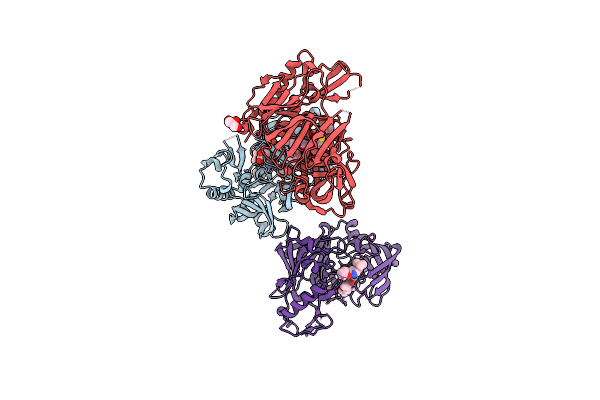
Crystal Structure Of Bace1 In Complex With A 2-Aminooxazoline 4-Azaxanthene Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-01-21 Classification: Hydrolase/Hydrolase inhibitor Ligands: 3LL, IOD, GOL |
 |
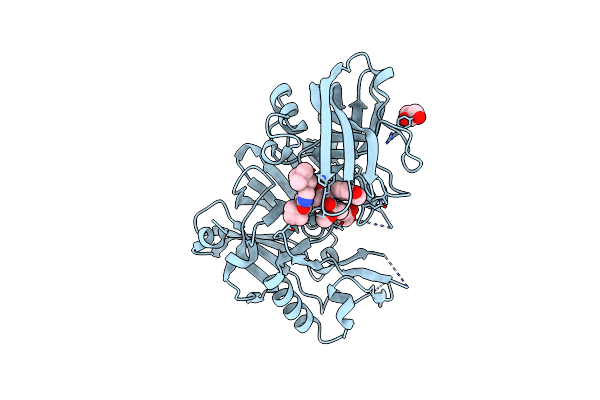
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-10-10 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 0K9, GOL |
 |
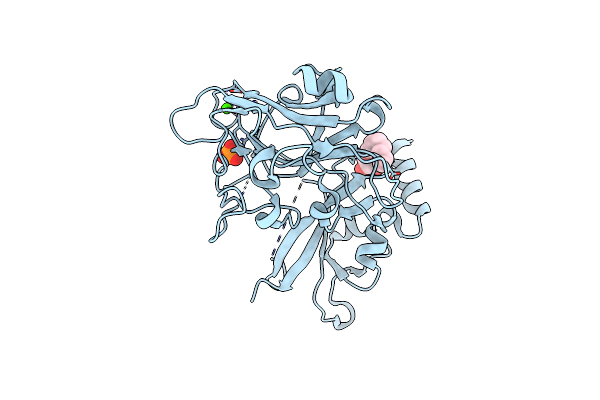
Structure Of Bace-1 (Beta-Secretase) In Complex With (2R)-N-((2S,3R)-1-(Benzo[D][1,3]Dioxol-5-Yl)-3-Hydroxy-4-((S)-6'-Neopentyl-3',4'-Dihydrospiro[Cyclobutane-1,2'-Pyrano[2,3-B]Pyridine]-4'-Ylamino)Butan-2-Yl)-2-Methoxypropanamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-04-18 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: IOD, GOL, 0KN |
 |
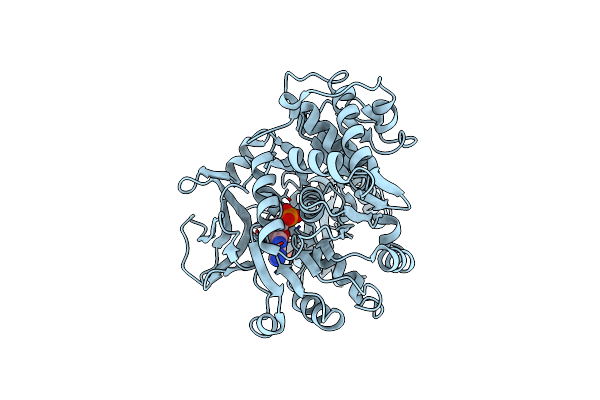
Crystal Structure Of The C-Terminal Extracellular Domain Of Mycobacterium Tuberculosis Embc
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-12-15 Classification: TRANSFERASE Ligands: AFO, CA, PO4 |
 |
Structure Of The U65C Mutant A-Riboswitch Aptamer From The Bacillus Subtilis Pbue Operon
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-01-19 Classification: RNA Ligands: MG, BR |
 |
Crystal Structure Of Dlta: Implications For The Reaction Mechanism Of Non-Ribosomal Peptide Synthetase (Nrps) Adenylation Domains
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2008-09-09 Classification: LIGASE Ligands: PO4, AMP |
 |
Crystal Structure Of Dlta: Implications For The Reaction Mechanism Of Non-Ribosomal Peptide Synthetase (Nrps) Adenylation Domains
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2008-09-09 Classification: LIGASE Ligands: AMP |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2002-09-11 Classification: LIGASE Ligands: SO4, AMP, DBH |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2002-09-11 Classification: LIGASE Ligands: SO4 |

