Search Count: 20
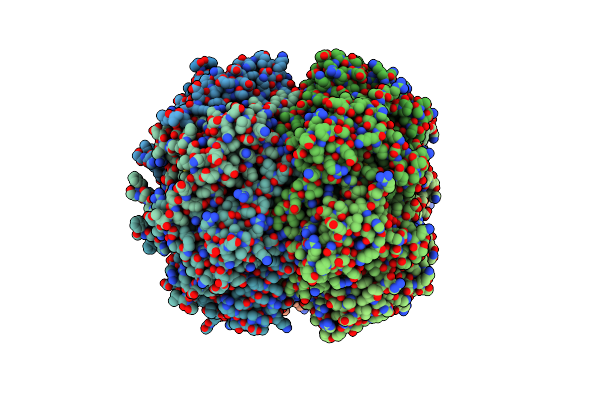 |
Crystal Structure Of The Active Form Of The Proteolytic Complex Clpp1 And Clpp2
Organism: Mycobacterium tuberculosis (strain cdc 1551 / oshkosh), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.07 Å Release Date: 2016-02-17 Classification: HYDROLASE |
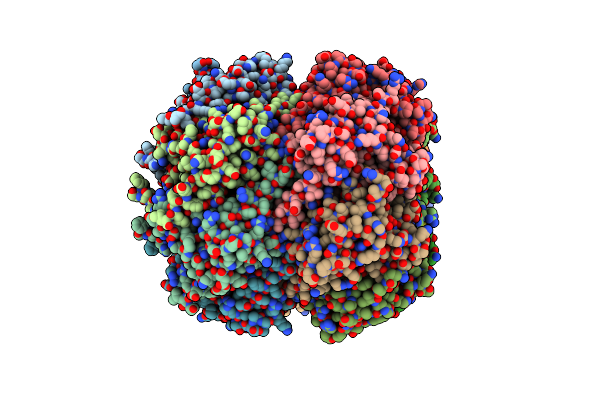 |
Crystal Structure Of The Active Form Of The Proteolytic Complex Clpp1 And Clpp2
Organism: Mycobacterium tuberculosis (strain cdc 1551 / oshkosh)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2016-02-17 Classification: HYDROLASE |
 |
Organism: Escherichia coli, Streptomyces hawaiiensis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-11-03 Classification: HYDROLASE/ANTIBIOTIC Ligands: MPD |
 |
Organism: Thermococcus onnurineus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-09-22 Classification: HYDROLASE Ligands: ADP, PE8, PE4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2010-07-21 Classification: HYDROLASE |
 |
Crystal Structure Of E. Coli Clpp With A Peptide Chloromethyl Ketone Covalently Bound At The Active Site
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-05-23 Classification: HYDROLASE Ligands: CMQ, PGE, GOL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2005-11-01 Classification: HYDROLASE |
 |
Crystallography And Mutagenesis Point To An Essential Role For The N-Terminus Of Human Mitochondrial Clpp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2005-03-22 Classification: HYDROLASE Ligands: DIO, EDO, GOL, FME |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2005-02-15 Classification: HYDROLASE |
 |
Atp-Dependent Clp Protease Atp-Binding Subunit Clpa/Atp-Dependent Clp Protease Adaptor Protein Clps
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2005-02-15 Classification: HYDROLASE Ligands: ZN, YBT, GOL, CL, Y1 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2005-02-15 Classification: CHAPERONE/PROTEIN BINDING Ligands: YBT, GOL, Y1 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2004-08-24 Classification: HYDROLASE Ligands: MG, ADP |
 |
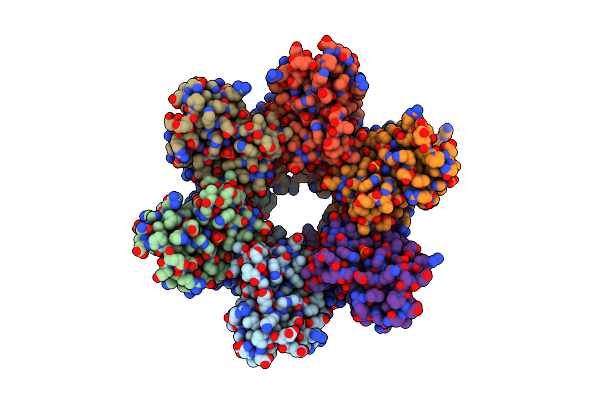
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-05-04 Classification: HYDROLASE |
 |
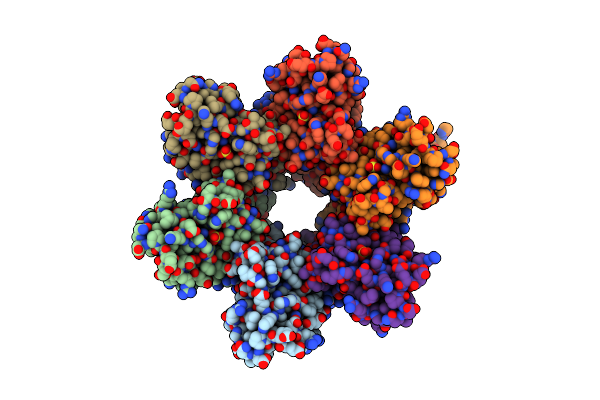
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2004-02-03 Classification: HYDROLASE Ligands: SO4 |
 |
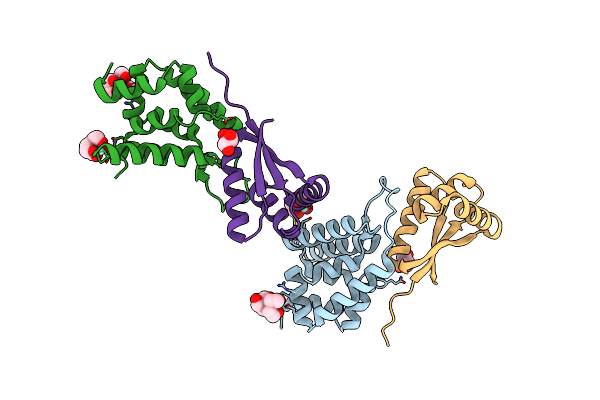
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2003-12-23 Classification: HYDROLASE Ligands: SO4 |
 |
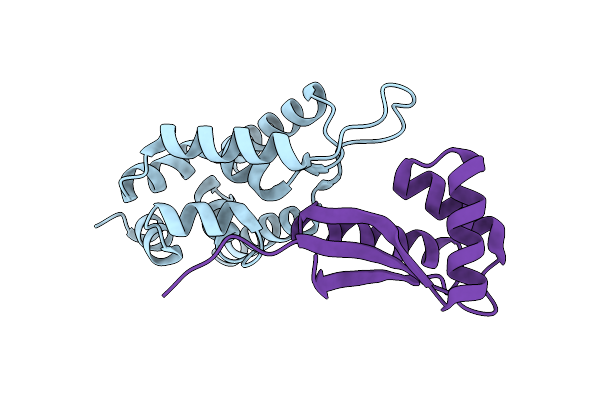
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2002-12-11 Classification: PROTEIN BINDING Ligands: YBT, GOL, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2002-12-11 Classification: PROTEIN BINDING |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2002-12-11 Classification: PROTEIN BINDING Ligands: ZN, YBT, GOL, CL |
 |
Crystal Structure Of Clpa, An Aaa+ Chaperone-Like Regulator Of Clpap Protease Implication To The Functional Difference Of Two Atpase Domains
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2002-09-27 Classification: HYDROLASE |
 |
Crystal Structure Of Clpa, An Hsp100 Chaperone And Regulator Of Clpap Protease: Structural Basis Of Differences In Function Of The Two Aaa+ Atpase Domains
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2002-09-27 Classification: HYDROLASE, LIGAND BINDING PROTEIN Ligands: MG, MET, PGE, ADP, IPA |

