Search Count: 38
 |
Organism: Malus domestica
Method: ELECTRON MICROSCOPY Resolution:3.00 Å Release Date: 2022-04-06 Classification: MEMBRANE PROTEIN |
 |
Organism: Malus domestica
Method: ELECTRON MICROSCOPY Resolution:2.70 Å Release Date: 2022-04-06 Classification: MEMBRANE PROTEIN |
 |
Organism: Malus domestica
Method: ELECTRON MICROSCOPY Resolution:2.70 Å Release Date: 2022-04-06 Classification: MEMBRANE PROTEIN |
 |
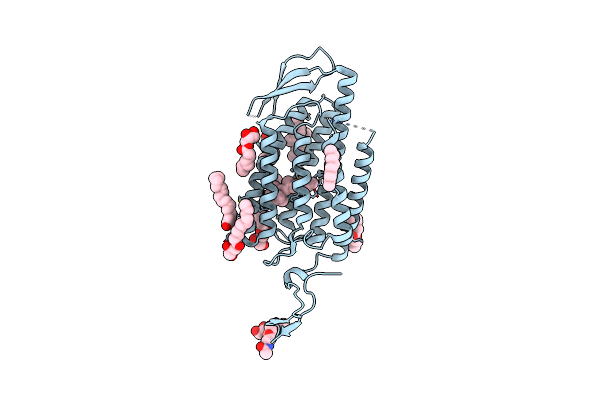
Organism: Thermus thermophilus hb8, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2021-04-14 Classification: TRANSPORT PROTEIN/IMMUNE SYSTEM |
 |
Time-Resolved Serial Femtosecond Crystallography Reveals Early Structural Changes In Channelrhodopsin: Dark State Structure
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-04-07 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
 |
Time-Resolved Serial Femtosecond Crystallography Reveals Early Structural Changes In Channelrhodopsin: 4 Ms Structure
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-04-07 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
 |
Time-Resolved Serial Femtosecond Crystallography Reveals Early Structural Changes In Channelrhodopsin: 1 Microsecond Structure
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-04-07 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
 |
Time-Resolved Serial Femtosecond Crystallography Reveals Early Structural Changes In Channelrhodopsin: 50 Microsecond Structure
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-04-07 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
 |
Time-Resolved Serial Femtosecond Crystallography Reveals Early Structural Changes In Channelrhodopsin: 250 Microsecond Structure
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-04-07 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
 |
Time-Resolved Serial Femtosecond Crystallography Reveals Early Structural Changes In Channelrhodopsin: 1 Ms Structure
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-04-07 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
 |
Crystal Structure Of Vibrio Cholerae Mate Transporter Vcmn In The Straight Form
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2019-01-16 Classification: TRANSPORT PROTEIN Ligands: OLC |
 |
Crystal Structure Of Vibrio Cholerae Mate Transporter Vcmn In The Bent Form
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-01-16 Classification: TRANSPORT PROTEIN Ligands: OLC |
 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2019-01-16 Classification: TRANSPORT PROTEIN Ligands: OLC |
 |
Crystal Structures Of The Tric Trimeric Intracellular Cation Channel Orthologue From Sulfolobus Solfataricus
Organism: Mus musculus, Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2017-01-11 Classification: IMMUNE SYSTEM/MEMBRANE PROTEIN Ligands: GOL, NA, PX4 |
 |
Crystal Structures Of The Tric Trimeric Intracellular Cation Channel Orthologue From Rhodobacter Sphaeroides
Organism: Rhodobacter sphaeroides (strain atcc 17023 / 2.4.1 / ncib 8253 / dsm 158)
Method: X-RAY DIFFRACTION Resolution:3.41 Å Release Date: 2017-01-11 Classification: MEMBRANE PROTEIN Ligands: PX4 |
 |
Structure Of A Nitrate/Nitrite Antiporter Nark In Nitrate-Bound Inward-Open State
Organism: Escherichia coli str. k12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-07-15 Classification: TRANSPORT PROTEIN Ligands: NO3, ZN, OLA, OLC |
 |
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2015-07-15 Classification: TRANSPORT PROTEIN Ligands: OLA, NI |
 |
Structure Of A Nitrate/Nitrite Antiporter Nark In Nitrate-Bound Occluded State
Organism: Escherichia coli str. k12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-07-15 Classification: TRANSPORT PROTEIN Ligands: NO3, OLA, OLC |
 |
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, OLA, ZN |
 |
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-04-08 Classification: MEMBRANE PROTEIN Ligands: RET, OLA, PEG |

