Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Crystal Structure Of Iron-Bound Human Ado C18S/C239S Variant Soaked In Hydralazine At 2.39 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: FE2, HLZ, GOL |
 |
Crystal Structure Of Cobalt-Bound Human Ado C18S/C239S Variant In Complex With Hydralazine At 1.89 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: OXIDOREDUCTASE Ligands: CO, HLZ, GOL, SO4 |
 |
The Crystal Structure Of Circular Mannose With Mutant H247F Of The Cellobiose 2-Epimerase From Caldicellulosiruptor Saccharolyticus
Organism: Caldicellulosiruptor saccharolyticus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-10-25 Classification: ISOMERASE Ligands: BMA |
 |
The Crystal Structure Of Linear Mannose With Mutant H247F Of The Cellobiose 2-Epimerase From Caldicellulosiruptor Saccharolyticus
Organism: Caldicellulosiruptor saccharolyticus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-10-25 Classification: ISOMERASE Ligands: DNO, BMA, EDO |
 |
Crystal Structure Of The Csce With Ligand To Have A Insight Into The Catalytic Mechanism
Organism: Caldicellulosiruptor saccharolyticus dsm 8903
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-12-01 Classification: PROTEIN BINDING Ligands: CL |
 |
Organism: Porphyromonas gingivalis tdc60
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-04-15 Classification: CELL ADHESION Ligands: SO4 |
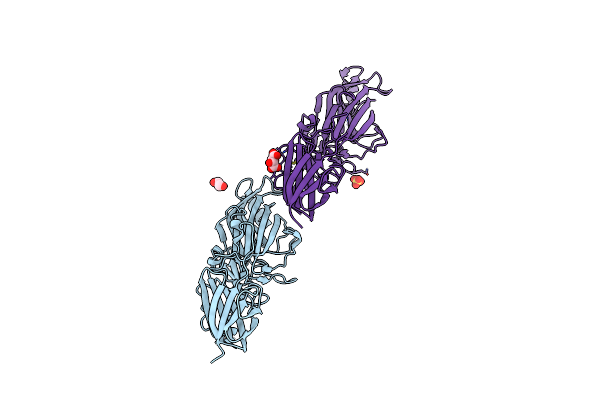 |
Organism: Porphyromonas gingivalis atcc 33277
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-04-15 Classification: CELL ADHESION Ligands: GOL, SO4, SRT |
 |
Organism: Porphyromonas gingivalis atcc 33277
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: CELL ADHESION |
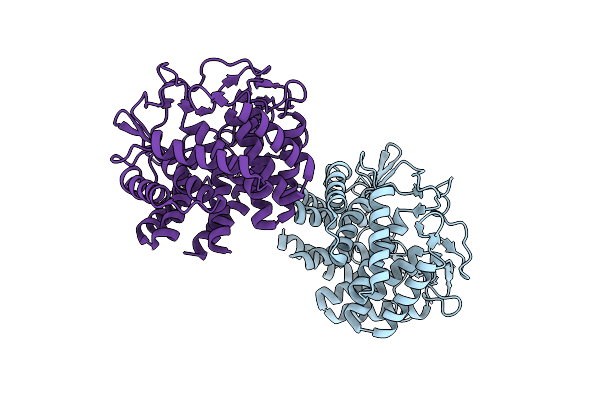 |
Organism: Caldibacillus thermoamylovorans
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-06-19 Classification: ISOMERASE |
 |
The Structure Of Cellobiose 2-Epimerase From Spirochaeta Thermophila Dsm 6192
Organism: Spirochaeta thermophila dsm 6192
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-04-10 Classification: ISOMERASE Ligands: EDO |
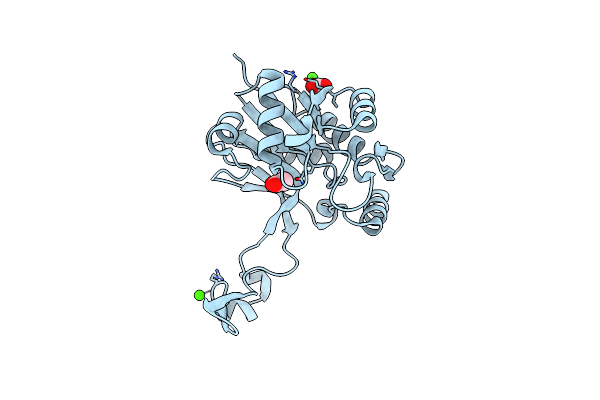 |
Organism: Neisseria meningitidis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-03-06 Classification: TRANSFERASE Ligands: ACT, CA |
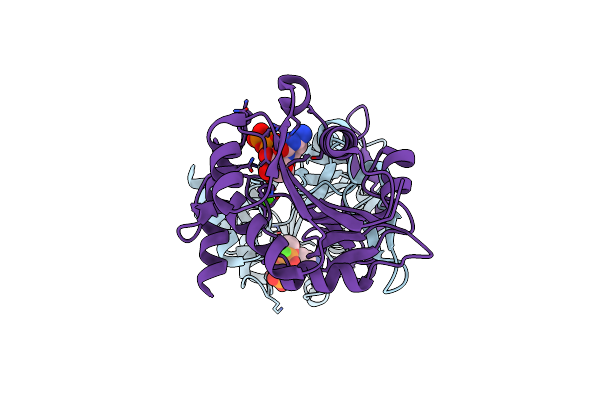 |
Organism: Neisseria meningitidis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-03-06 Classification: TRANSFERASE Ligands: CTP, CA |
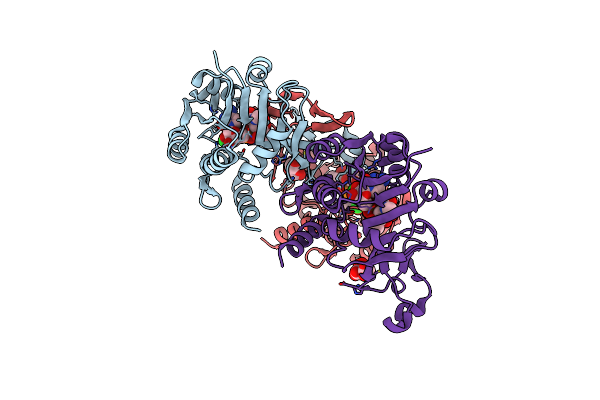 |
N. Meningitidis Cmp-Sialic Acid Synthetase In The Presence Of Cmp And Neu5Ac2En
Organism: Neisseria meningitidis
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2019-03-06 Classification: TRANSFERASE Ligands: C5P, CL, DAN, FLC |
 |
N. Meningitidis Cmp-Sialic Acid Synthetase In The Presence Of Cmp-Sialic Acid And Ca2+
Organism: Neisseria meningitidis
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2019-03-06 Classification: TRANSFERASE Ligands: NCC, CA, GOL |
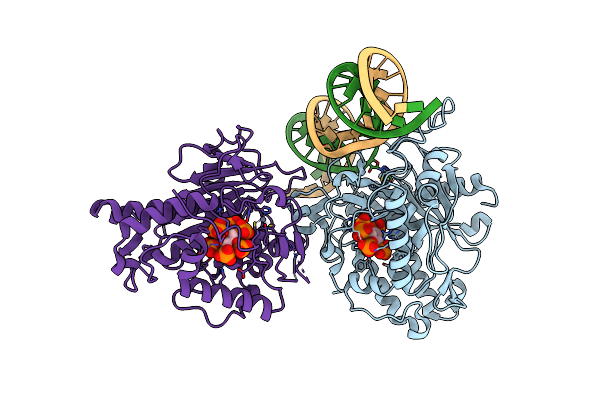 |
Human Adar2D E488Y Mutant Complexed With Dsrna Containing An Abasic Site Opposite The Edited Base
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2019-02-20 Classification: HYDROLASE/RNA Ligands: IHP, ZN |
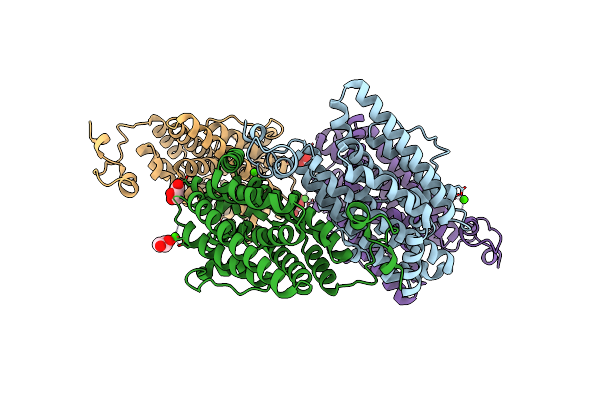 |
Crystal Structure Of The Class Ie Ribonucleotide Reductase Beta Subunit From Aerococcus Urinae In Unactivated Form
Organism: Aerococcus urinae (strain acs-120-v-col10a)
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2018-09-19 Classification: OXIDOREDUCTASE Ligands: CA, GOL |
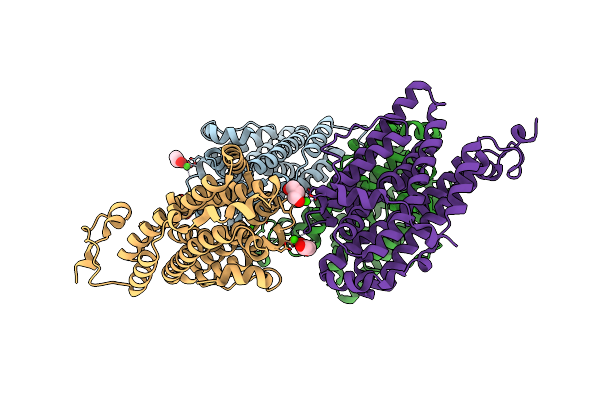 |
Crystal Structure Of The Class Ie Ribonucleotide Reductase Beta Subunit From Aerococcus Urinae In Activated Form
Organism: Aerococcus urinae (strain acs-120-v-col10a)
Method: X-RAY DIFFRACTION Release Date: 2018-09-19 Classification: OXIDOREDUCTASE Ligands: CA, GOL |
 |
Crystal Structure Of The Flavoprotein Nrdi From Aerococcus Urinae In Oxidized Form
Organism: Aerococcus urinae
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-09-19 Classification: FLAVOPROTEIN Ligands: FMN |
 |
Crystal Structure Of The Class Ie Ribonucleotide Reductase Beta Subunit From Aerococcus Urinae In Activated Form With Thiocyanate Bound
Organism: Aerococcus urinae (strain acs-120-v-col10a)
Method: X-RAY DIFFRACTION Release Date: 2018-09-19 Classification: OXIDOREDUCTASE Ligands: CA, GOL, SCN |
 |
Human Adenosine Deaminase Acting On Dsrna (Adar2) Mutant E488Q Bound To Dsrna Sequence Derived From S. Cerevisiae Bdf2 Gene
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2016-04-13 Classification: HYDROLASE/RNA Ligands: IHP, ZN |