Search Count: 188
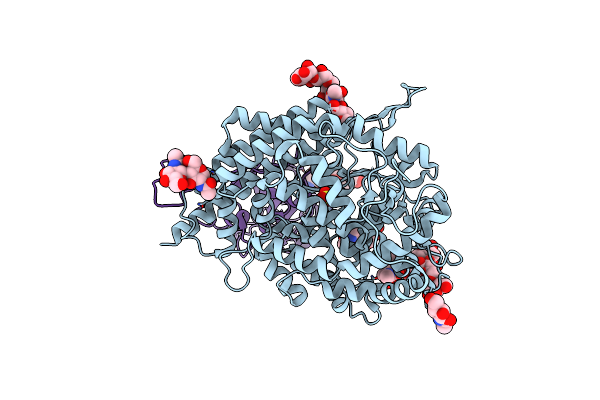 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2023-12-27 Classification: VIRAL PROTEIN/PROTEIN BINDING Ligands: NAG, ZN, SO4 |
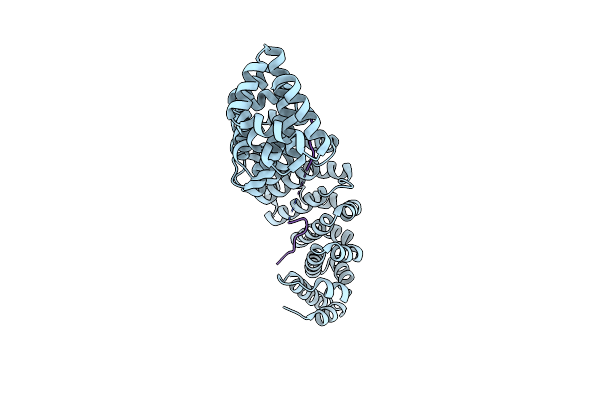 |
Crystal Structure Of Importin-Alpha1 Bound To The Hif-1Alpha Nuclear Localization Signal (Wild-Type)
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-02-22 Classification: TRANSPORT PROTEIN |
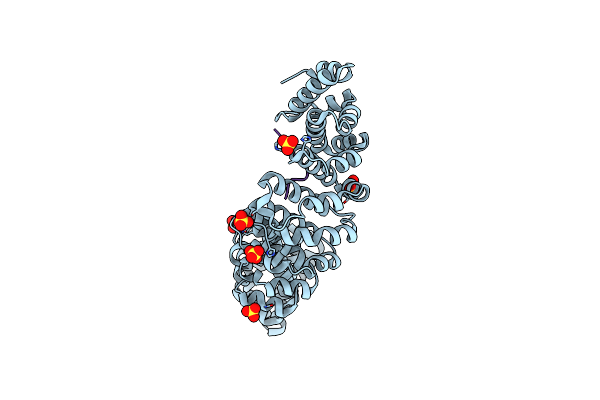 |
Crystal Structure Of Importin-Alpha1 Bound To The Hif-1Alpha Nuclear Localization Signal (Delta 724-751)
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-02-22 Classification: TRANSPORT PROTEIN Ligands: SO4 |
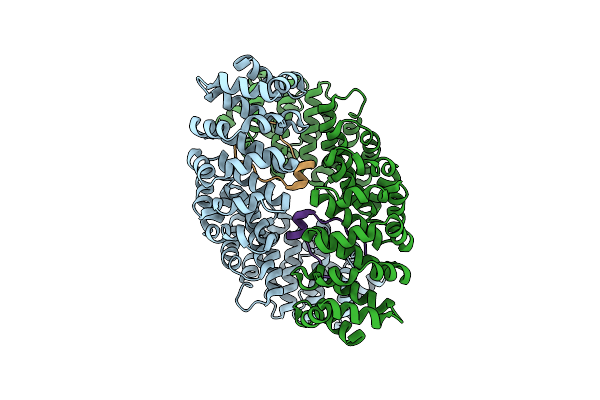 |
Crystal Structure Of Importin-Alpha3 Bound To The 53Bp1 Nuclear Localization Signal
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-12-21 Classification: TRANSPORT PROTEIN |
 |
Organism: Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-07-20 Classification: IMMUNE SYSTEM Ligands: SO4, EDO |
 |
Cryo-Em Structure Of The Sars-Cov-2 Spike Protein (2-Up Rbd) Bound To Neutralizing Nanobodies P86
Organism: Severe acute respiratory syndrome coronavirus 2, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2022-07-20 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Time-Resolved Serial Femtosecond Crystallography Structure Of Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3: Resting State Structure With Bromide Ion
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-02-16 Classification: MEMBRANE PROTEIN Ligands: RET, HEX, D10, BR |
 |
Time-Resolved Serial Femtosecond Crystallography Structure Of Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3 : Structure Obtained 1 Msec After Photoexcitation With Bromide Ion
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-02-16 Classification: MEMBRANE PROTEIN Ligands: RET, HEX, D10, BR |
 |
Anion Free Form Of Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3, Structure Determined By Serial Femtosecond Crystallography At Sacla
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-02-16 Classification: MEMBRANE PROTEIN Ligands: RET, HEX, DD9, C14, R16, OCT, CL |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-09-08 Classification: VIRAL PROTEIN |
 |
Ambient Temperature Structure Of Bifidobacterium Longum Phosphoketolase With Thiamine Diphosphate
Organism: Bifidobacterium longum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-06-02 Classification: LYASE Ligands: TPP, CA, LMR, MLA, SIN |
 |
Ambient Temperature Structure Of Bifidobacgterium Longum Phosphoketolase With Thiamine Diphosphate And Phosphoenol Pyuruvate
Organism: Bifidobacterium longum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-06-02 Classification: LYASE Ligands: PEP, TPP, CA |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2021-06-02 Classification: VIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-06-02 Classification: VIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-06-02 Classification: VIRAL PROTEIN |
 |
 |
 |
 |
Structure Of Sars-Cov-2 Spike Receptor-Binding Domain Complexed With High Affinity Ace2 Mutant 3N39
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2020-12-23 Classification: VIRAL PROTEIN Ligands: NAG, ZN, SO4 |
 |
Crystal Structure Of Human Fer Sh2 Domain Bound To A Phosphopeptide (Depyenvd)
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2019-09-04 Classification: TRANSFERASE/SIGNALING PROTEIN |

