Search Count: 38
 |
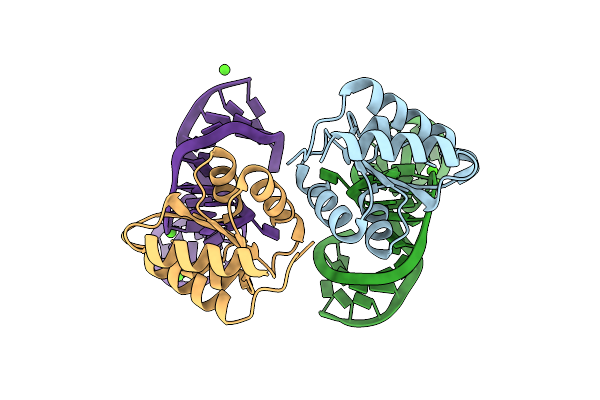
Crystal Structure Of The L7Ae Derivative Protein Ls4 In Complex With Its Co-Evolved Target Cs1 Rna
Organism: Archaeoglobus fulgidus (strain atcc 49558 / dsm 4304 / jcm 9628 / nbrc 100126 / vc-16), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-04-02 Classification: RNA BINDING PROTEIN Ligands: MG |
 |
Crystal Structure Of The L7Ae Derivative Protein Ls12 In Complex With Its Co-Evolved Target Cs2 Rna
Organism: Archaeoglobus fulgidus (strain atcc 49558 / dsm 4304 / jcm 9628 / nbrc 100126 / vc-16), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.12 Å Release Date: 2025-04-02 Classification: RNA BINDING PROTEIN Ligands: CA |
 |
Organism: Archaeoglobus fulgidus dsm 4304
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-04-02 Classification: RNA BINDING PROTEIN |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: TOXIN |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: TOXIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2022-07-27 Classification: TRANSFERASE Ligands: 3UI, NO3 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-01-05 Classification: OXIDOREDUCTASE Ligands: HEM, MYT, CHD, GOL |
 |
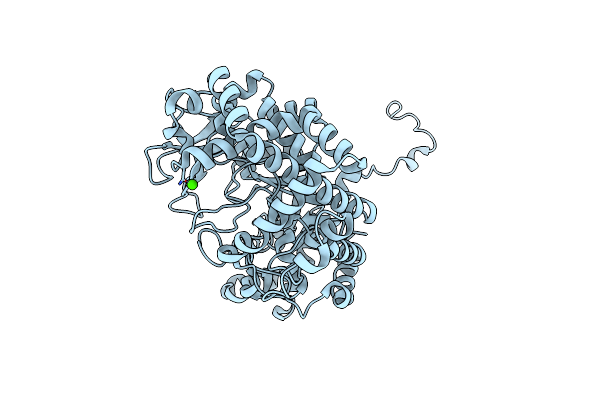
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: GOL, CA, MG |
 |
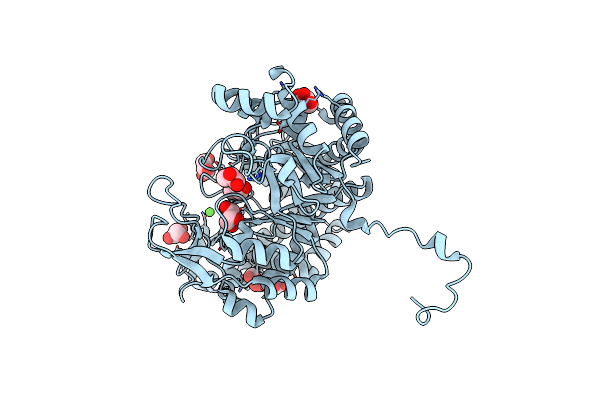
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: CA |
 |
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: CA, GOL |
 |
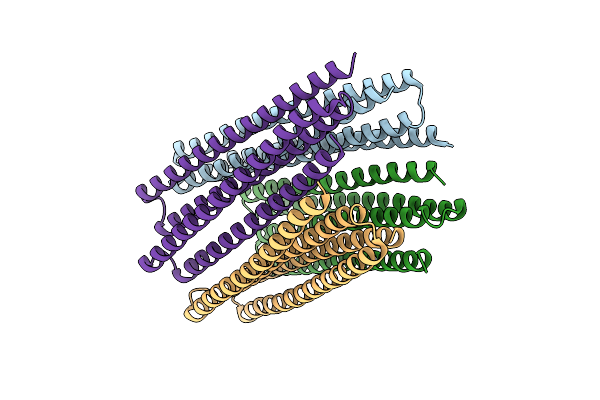
Cryo-Em Structure Of A De Novo Designed 16-Helix Transmembrane Nanopore, Tmhc8_R.
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-08-19 Classification: DE NOVO PROTEIN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2020-06-24 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-04-29 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-04-29 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-03-18 Classification: DE NOVO PROTEIN |
 |
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2016-12-28 Classification: CELL ADHESION/IMMUNE SYSTEM Ligands: GOL, PO4, CL |
 |
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2016-12-28 Classification: CELL ADHESION/IMMUNE SYSTEM Ligands: CA |
 |
Cytoplasmic Domain Of Inward Rectifier Potassium Channel Kir3.2 In Complex With Cadmium
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.56 Å Release Date: 2011-10-05 Classification: TRANSPORT PROTEIN Ligands: CD, MG, EOH |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2011-03-23 Classification: HYDROLASE Ligands: CL, NA, ACT |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2011-03-23 Classification: HYDROLASE Ligands: ACT, NA, CL |

