Search Count: 17
 |
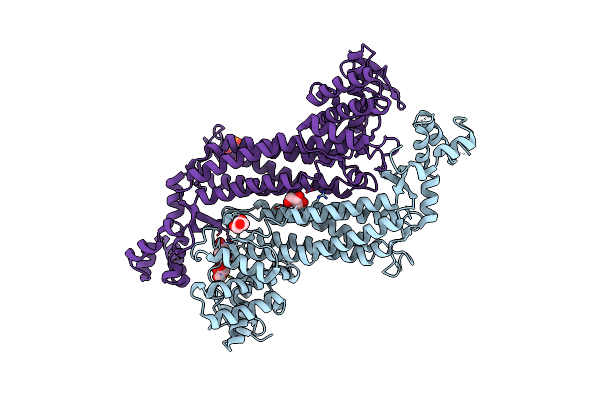
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-11-09 Classification: LYASE Ligands: PO4, MG, ACT, EDO, PGE, SEP |
 |
Crystal Structure Of Recombinant Human Fumarase In Complex With D-2-Amino-3-Phosphono-Propionic Acid
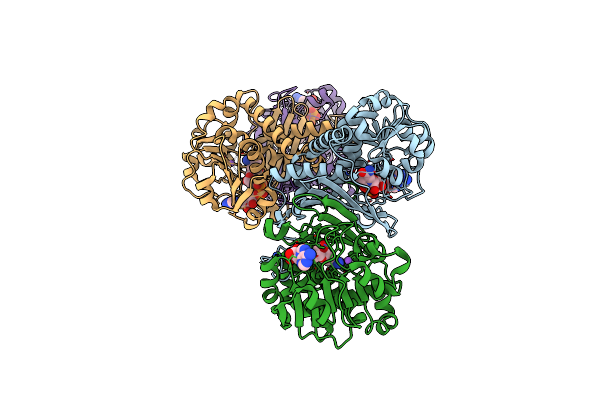
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-02-23 Classification: LYASE Ligands: APO, GOL |
 |
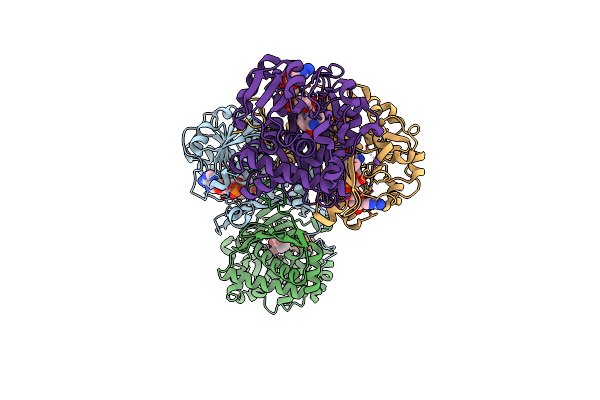
Structural Basis For Recognition Of L-Lysine, L-Ornithine, And L-2,4-Diamino Butyric Acid By Lysine Cyclodeaminase
Organism: Streptomyces pristinaespiralis
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2018-05-02 Classification: LYASE |
 |
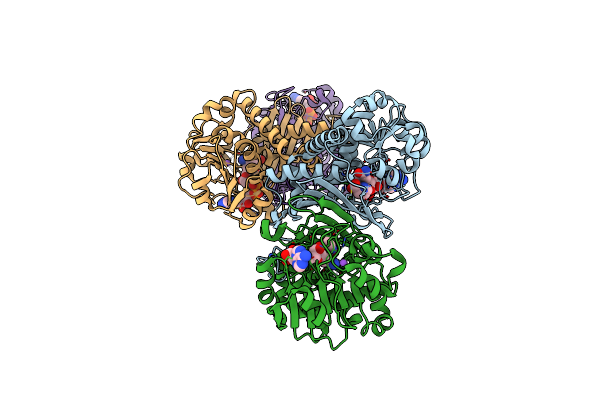
Structural Basis For Recognition Of L-Lysine, L-Ornithine, And L-2,4-Diamino Butyric Acid By Lysine Cyclodeaminase
Organism: Streptomyces pristinaespiralis
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2018-05-02 Classification: LYASE |
 |
Structural Basis For Recognition Of L-Lysine, L-Ornithine, And L-2,4-Diamino Butyric Acid By Lysine Cyclodeaminase
Organism: Streptomyces pristinaespiralis
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2018-05-02 Classification: LYASE |
 |
Structural Basis For Recognition Of L-Lysine, L-Ornithine, And L-2,4-Diamino Butyric Acid By Lysine Cyclodeaminase
Organism: Streptomyces pristinaespiralis
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2018-05-02 Classification: LYASE |
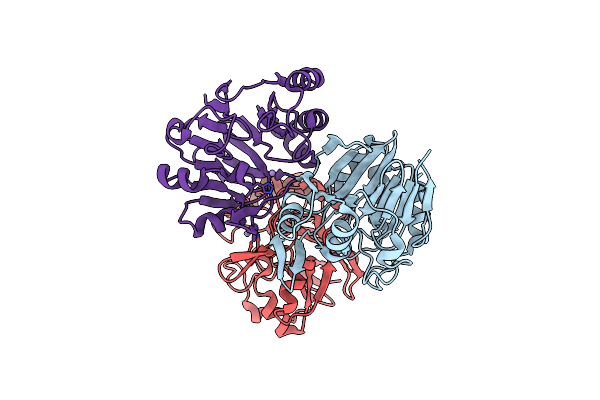 |
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:3.42 Å Release Date: 2016-02-17 Classification: HYDROLASE Ligands: MN |
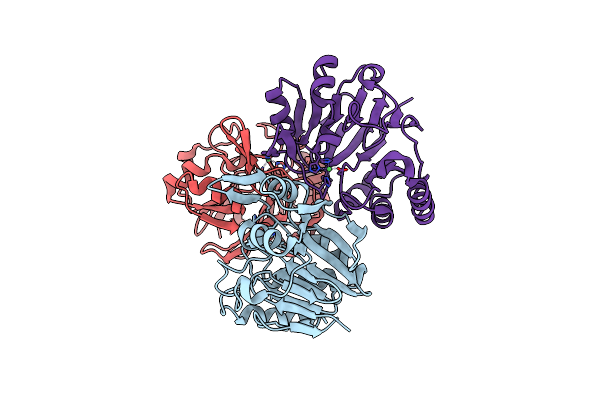 |
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2016-02-17 Classification: HYDROLASE Ligands: NI |
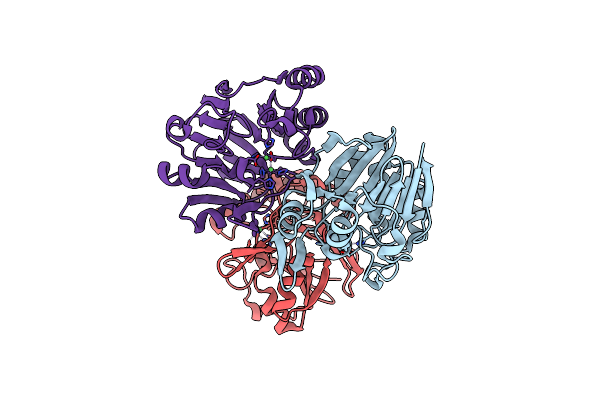 |
Crystal Structure Of Metallo-Beta-Lactamase In Complex With Nickel From Thermotoga Maritima
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2016-02-17 Classification: HYDROLASE Ligands: NI |
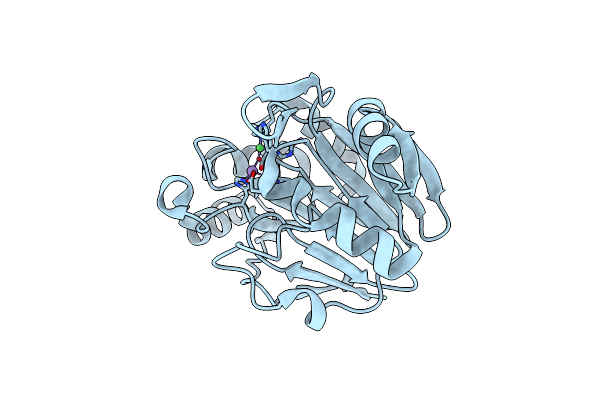 |
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2016-02-17 Classification: HYDROLASE Ligands: MN, NI |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-07-15 Classification: CELL CYCLE Ligands: GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-07-15 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2015-07-15 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-02-26 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of Pyridoxal Biosynthesis Lyase Pdxs From Pyrococcus Horikoshii
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-11-14 Classification: LYASE |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2012-11-14 Classification: LYASE Ligands: R5P |
 |
Organism: Bombyx mori
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2001-03-21 Classification: HYDROLASE |

