Search Count: 20
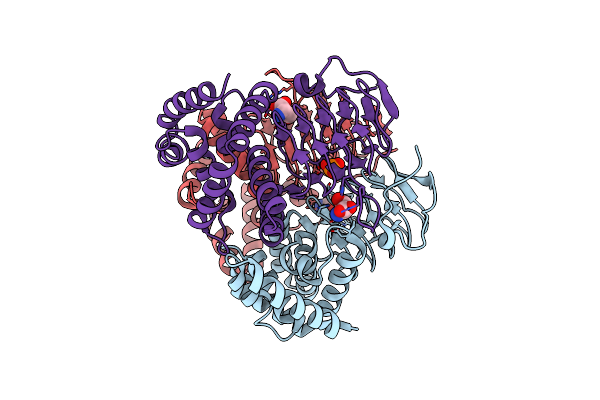 |
Crystal Structure Of Serine Acetyltransferase From Salmonella Typhimurium Complexed With Serine
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-11-22 Classification: TRANSFERASE Ligands: SER, PO4 |
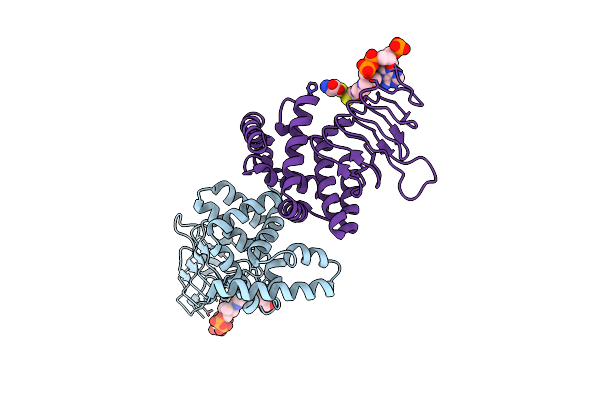 |
Crystal Structure Of Serine Acetyltransferase From Salmonella Typhimurium Complexed With Coa
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-11-22 Classification: TRANSFERASE Ligands: CYS, COA |
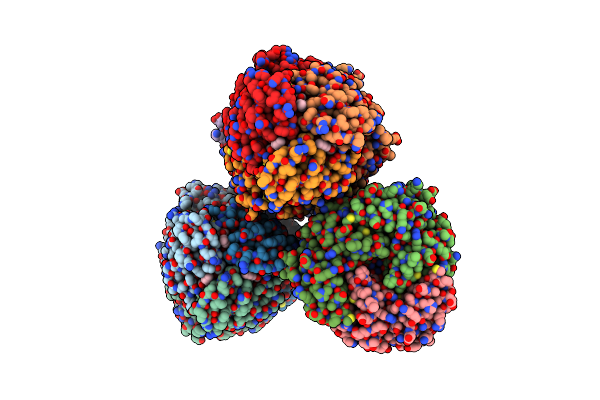 |
Crystal Structure Of Serine Acetyltransferase From Salmonella Typhimurium Complexed With Butyl Gallate
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-11-22 Classification: TRANSFERASE Ligands: CYS, NF0, PO4 |
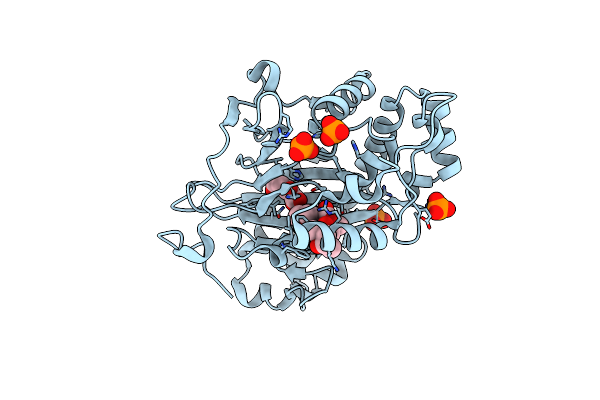 |
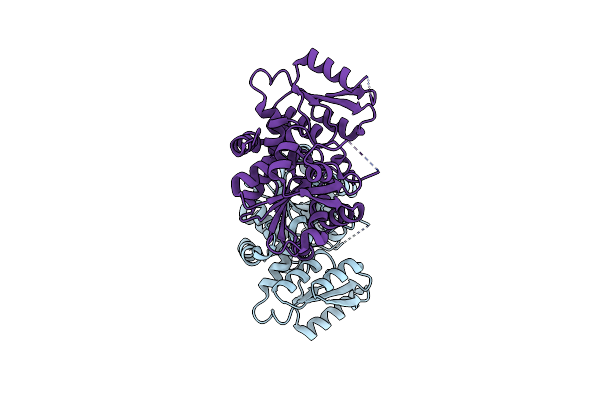
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-02-16 Classification: BIOSYNTHETIC PROTEIN Ligands: PO4, AKG, MPD, J5X |
 |
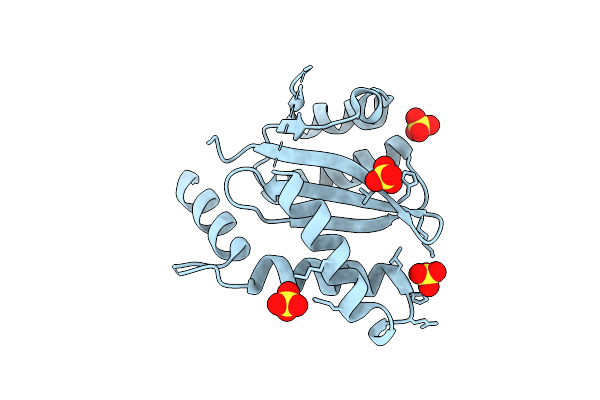
Organism: Dictyostelium discoideum
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2020-12-09 Classification: ISOMERASE |
 |
Crystal Structure Of Hiv-1 Integrase Catalytic Core Domain (A128T/K173Q/F185K)
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-07-01 Classification: VIRAL PROTEIN Ligands: SO4, ARS |
 |
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: GA4, AKG, GOL, SO4 |
 |
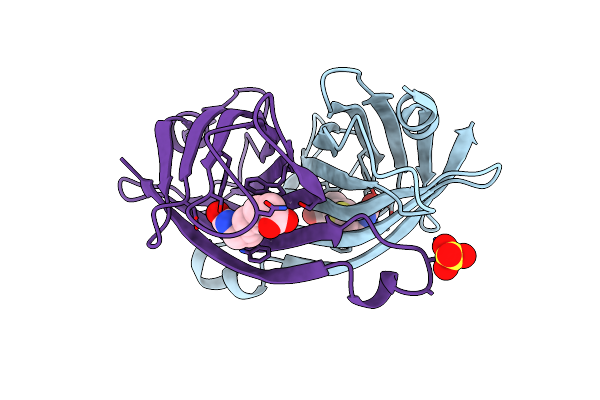
Organism: Oryza sativa subsp. indica
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: AKG, IAC, GOL |
 |
Crystal Structure Of The Core Streptavidin Mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) Complexed With Biotin At 1.3 A Resolution
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2015-02-18 Classification: BIOTIN BINDING PROTEIN Ligands: BTN, SO4 |
 |
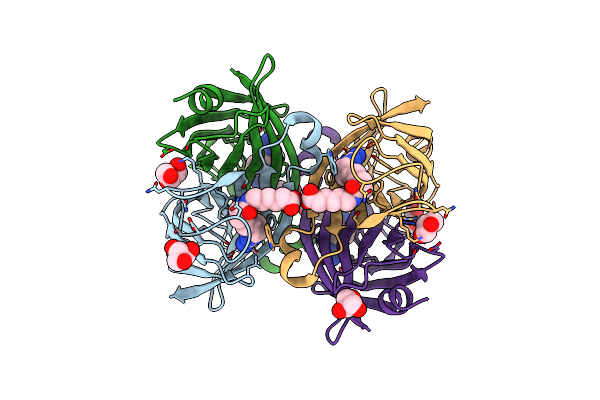
Crystal Structure Of The Core Streptavidin Mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) Complexed With Biotin Long Tail (Btntail) At 1.5 A Resolution
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-02-18 Classification: BIOTIN BINDING PROTEIN Ligands: ZOE, GOL, CD, P6G |
 |
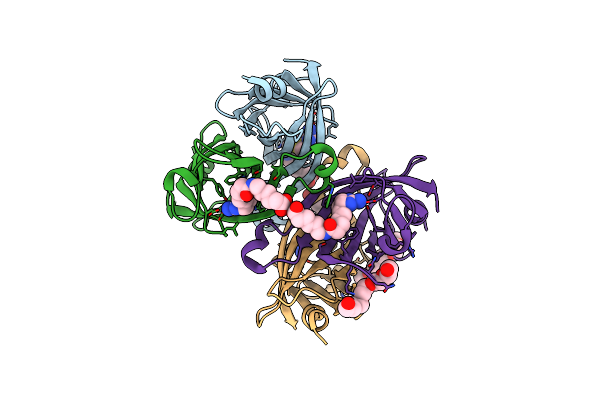
Crystal Structure Of The Core Streptavidin Mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) Complexed With Iminobiotin Long Tail (Imntail) At 1.2 A Resolution
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2015-02-18 Classification: BIOTIN BINDING PROTEIN Ligands: ZOF, GOL |
 |
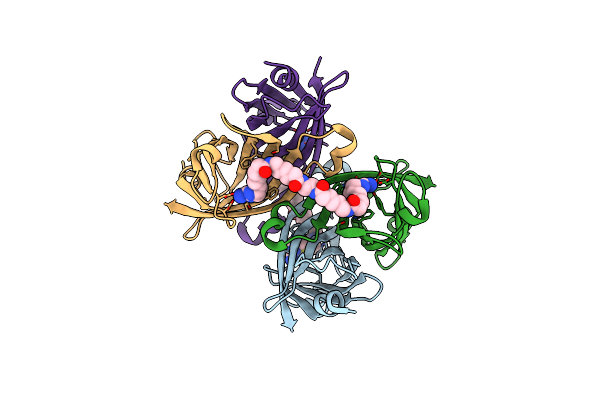
Crystal Structure Of The Core Streptavidin Mutant V212 (Y22S/N23D/S27D/S45N/Y83S/R84K/E101D/R103K/E116N) Complexed With Iminobiotin Long Tail (Imntail) At 1.7 A Resolution
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-02-18 Classification: BIOTIN BINDING PROTEIN Ligands: ZOF, P6G |
 |
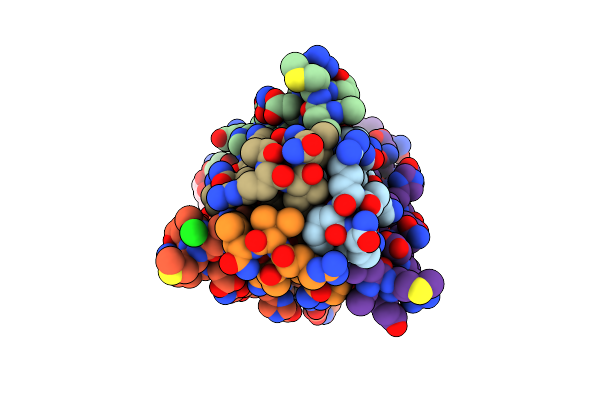
Crystal Structure Of The Core Streptavidin Mutant V212 (Y22S/N23D/S27D/S45N/Y83S/R84K/E101D/R103K/E116N) Complexed With Bis Iminobiotin Long Tail (Bis-Imntail) At 1.3 A Resolution
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2015-01-21 Classification: BIOTIN BINDING PROTEIN Ligands: ZOF, EDN |
 |
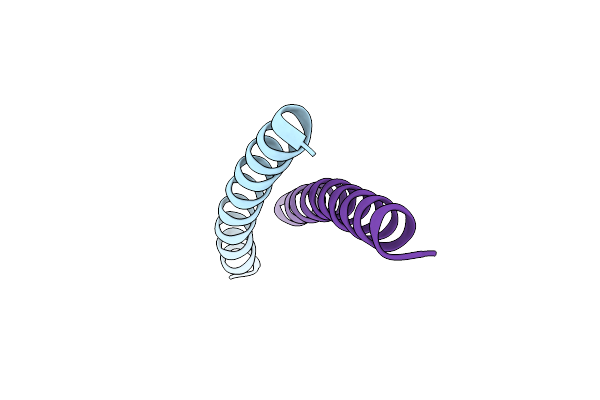
Crystal Structure Of The Complex Between Gp41 Fragments N36 And C34 Mutant N126K/E137Q
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-05-19 Classification: MEMBRANE PROTEIN Ligands: MPD, CL |
 |
Crystal Structure Of The Complex Between Gp41 Fragment N36 And Fusion Inhibitor C34/S138A
Method: X-RAY DIFFRACTION
Resolution:2.20 Å Release Date: 2009-08-04 Classification: VIRAL PROTEIN/INHIBITOR |
 |
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-11-25 Classification: HYDROLASE RECEPTOR Ligands: GA4, MPD, NO3, PO4 |
 |
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-11-25 Classification: HYDROLASE RECEPTOR Ligands: MPD, NO3, GA3, PO4 |
 |
Crystal Structure Of The Complex Between Gp41 Fragment N36 And Fusion Inhibitor Sc34Ek
Method: X-RAY DIFFRACTION
Resolution:2.10 Å Release Date: 2008-06-03 Classification: VIRAL PROTEIN/INHIBITOR Ligands: ACY, SO4 |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2005-03-08 Classification: TRANSFERASE Ligands: MG, SO4 |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2005-03-08 Classification: TRANSFERASE Ligands: MG, SO4, PEP |

