Search Count: 24
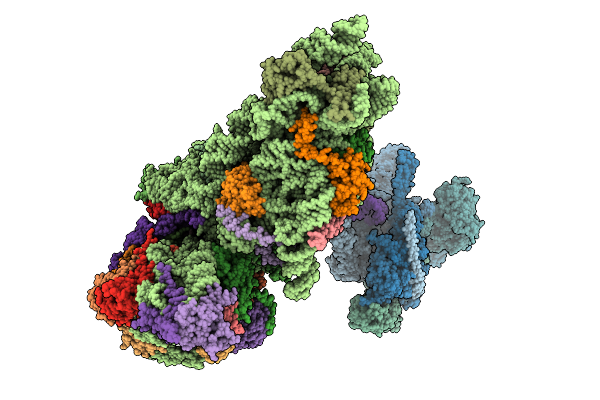 |
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG |
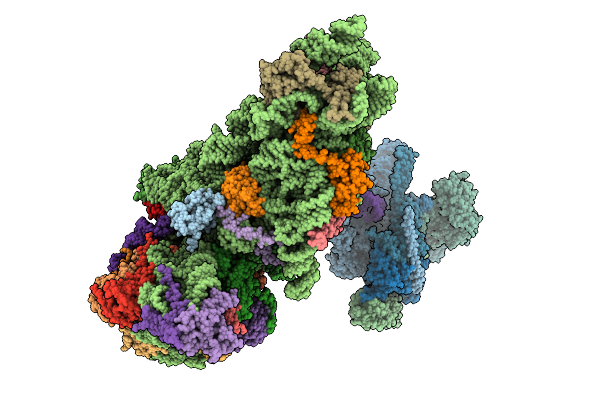 |
Structure Of The Human 40S Ribosome Complexed With Hcv Ires, Eif1A And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG |
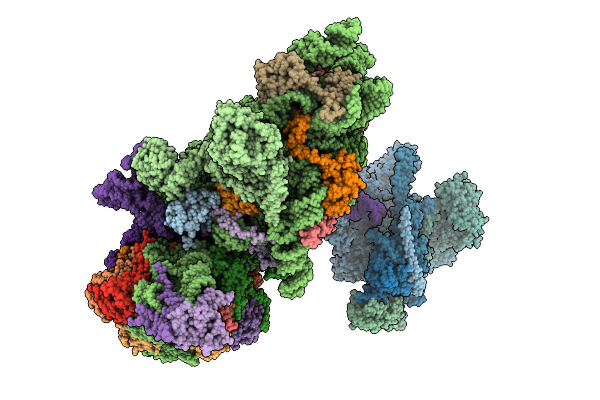 |
Structure Of The Hcv Ires-Dependent Pre-48S Translation Initiation Complex With Eif1A, Eif5B, And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, GTP, ZN |
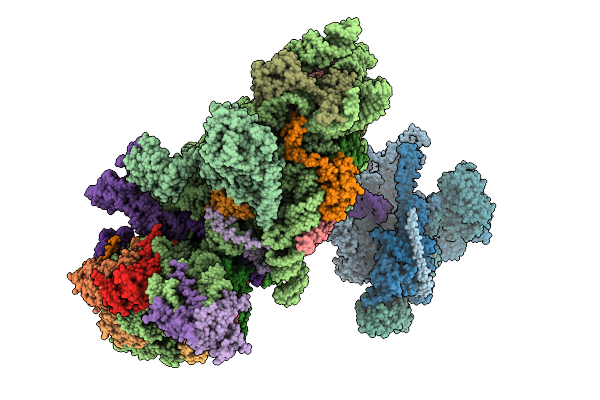 |
Structure Of The Hcv Ires-Dependent 48S Translation Initiation Complex With Eif5B And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG, GTP |
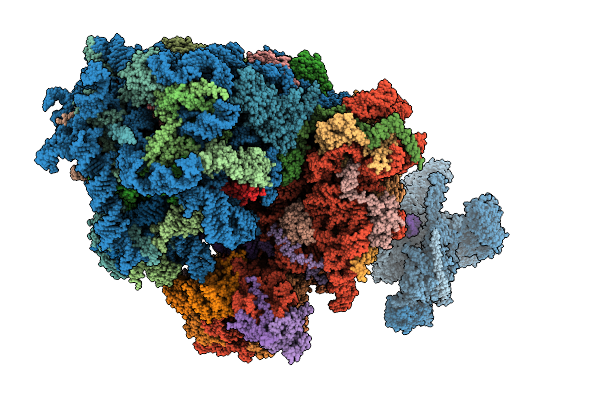 |
Cryo-Em Structure Of The Hcv Ires-Dependently Initiated Cmv-Stalled 80S Ribosome (Non-Rotated State) In Complexed With Eif3
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, ZN |
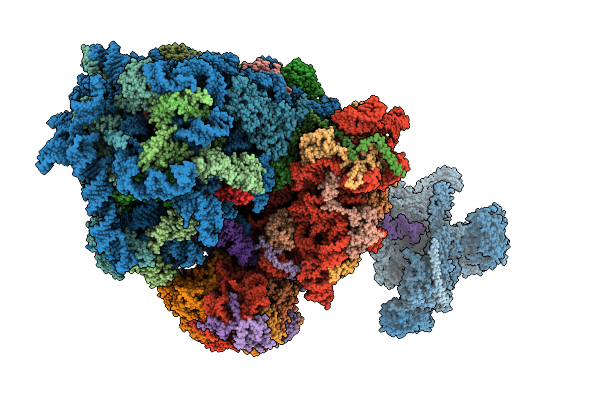 |
Cryo-Em Structure Of The Hcv Ires-Dependently Initiated Cmv-Stalled 80S Ribosome (Rotated State) In Complexed With Eif3
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: FLUORESCENT PROTEIN Ligands: ABU |
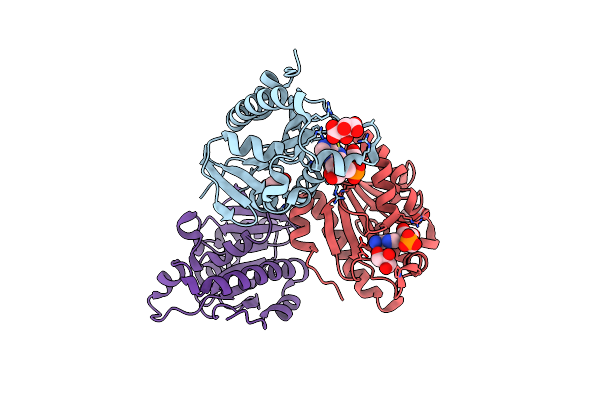 |
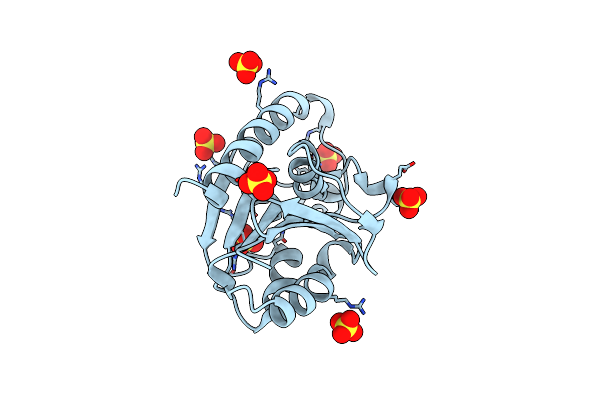
Crystal Structure Of Thermus Thermophilus Peptidyl-Trna Hydrolase In Complex With Adenosine 5'-Monophosphate
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Release Date: 2024-11-20 Classification: TRANSLATION Ligands: AMP, TLA, GOL |
 |
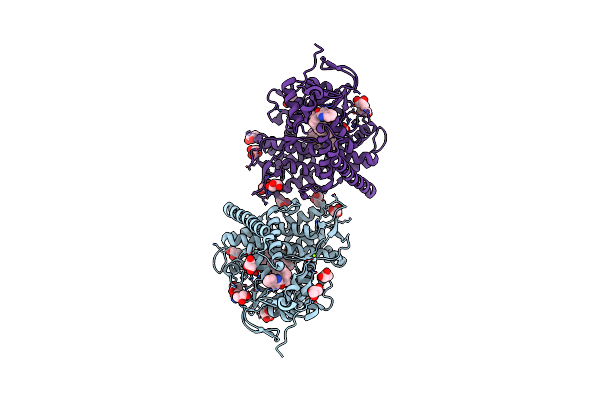
Crystal Structure Of Thermus Thermophilus Peptidyl-Trna Hydrolase C-Terminal 16 Amino Acid Deletion Mutant
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Release Date: 2024-11-20 Classification: TRANSLATION Ligands: SO4 |
 |
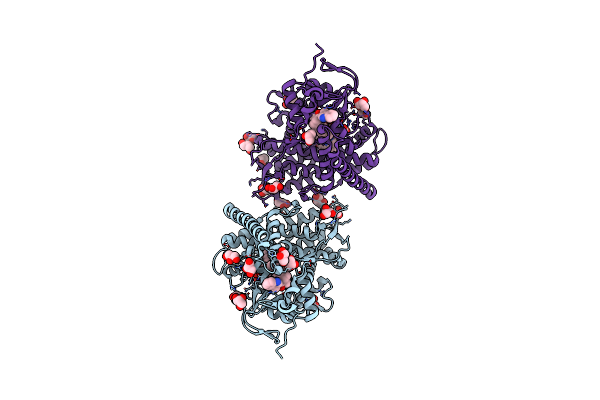
Near-Atomic Resolution Structure Of The Cyp102A1 Haem Domain With N-Abietoyl-L-Tryptophan
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: HEM, WAA, GOL, TRS, MG |
 |
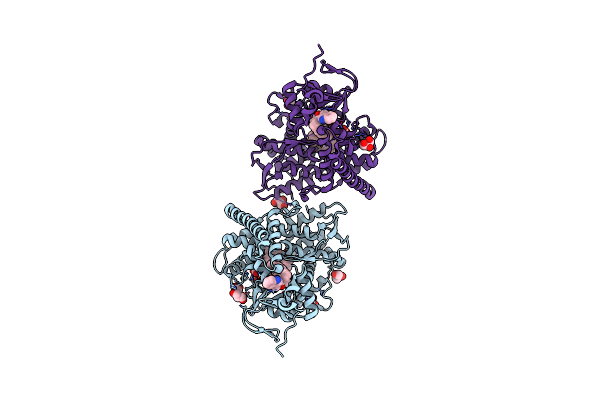
Structure Of The Oxomolybdenum Mesoporphyrin Ix-Reconstituted Cyp102A1 Haem Domain With N-Abietoyl-L-Tryptophan
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: MI9, WAA, GOL, TRS |
 |
Structure Of The Chromium Protoporphyrin Ix-Reconstituted Cyp102A1 Haem Domain With N-Abietoyl-L-Tryptophan
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: BW9, WAA, GOL |
 |
Structure Of The Cyp102A1 Haem Domain With N-(S)-Ibuprofenoyl-L-Phenylalanine
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: HEM, BWX, DMS, GOL |
 |
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: HEM, DMS, C5R, GOL |
 |
Structure Of The Cobalt Protoporphyrin Ix-Reconstituted Cyp102A1 Haem Domain With N-Abietoyl-L-Tryptophan
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: WAA, GOL, COH, DMS, TRS |
 |
Structure Of The Manganese Protoporphyrin Ix-Reconstituted Cyp102A1 Haem Domain With N-Abietoyl-L-Tryptophan In Complex With Pyridine
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: 0PY, WAA, MNH, GOL |
 |
Structure Of The Rhodium Mesoporphyrin Ix-Reconstituted Cyp102A1 Haem Domain With N-Abietoyl-L-Tryptophan
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: WAA, CV0, DMS |
 |
Structure Of The Cyp102A1 Haem Domain With N-Enanthyl-L-Prolyl-L-Phenylalanine
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: HEM, DMS, D0L, GOL |
 |
Structure Of The Carbonylruthenium Mesoporphyrin Ix-Reconstituted Cyp102A1 Haem Domain With N-Abietoyl-L-Tryptophan
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: RUR, CMO, GOL, WAA |
 |
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Release Date: 2018-09-26 Classification: HYDROLASE Ligands: FLC, MPD |

