Search Count: 22
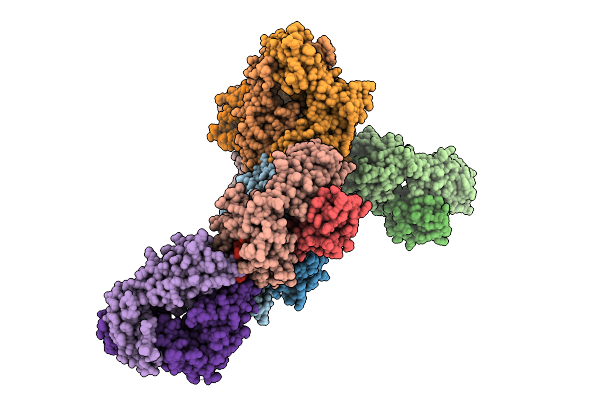 |
Human T Cell Receptor (Trav24*01/Trbv27*01) In Complex With Hla-C*1202 And Iy11 Peptide
Organism: Homo sapiens, Human immunodeficiency virus type 1 (z2/cdc-z34 isolate)
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: IMMUNE SYSTEM |
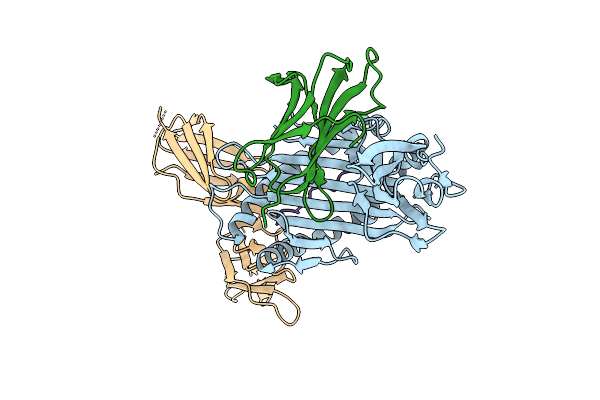 |
Killer Cell Immunoglobulin-Like Receptor 2Dl2 In Complex With Hla-C*1202 And Iy10 Peptide
Organism: Homo sapiens, Human immunodeficiency virus type 1 (z2/cdc-z34 isolate)
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: IMMUNE SYSTEM |
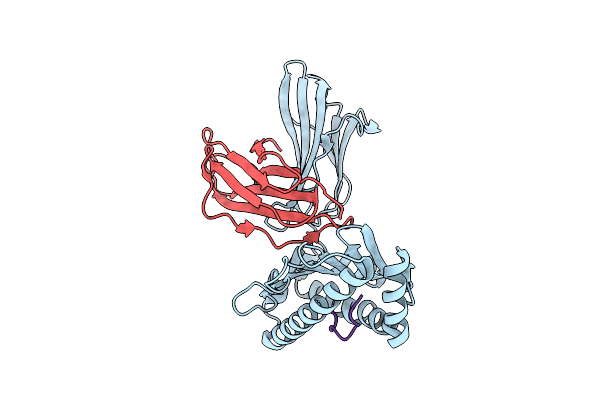 |
Organism: Homo sapiens, Human immunodeficiency virus type 1 (z2/cdc-z34 isolate)
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Human immunodeficiency virus type 1 (z2/cdc-z34 isolate)
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: IMMUNE SYSTEM |
 |
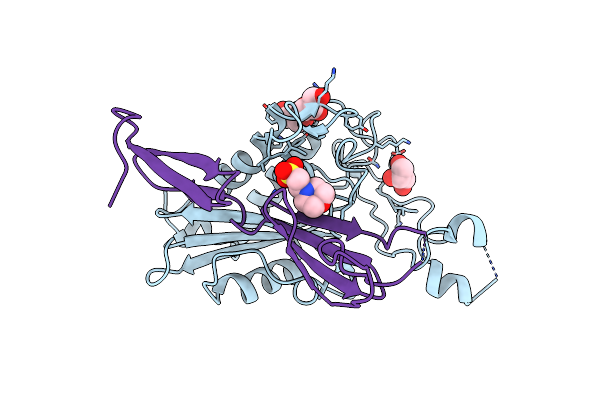
Structural And Functional Basis For Hla-G Isoform Recognition Of Immune Checkpoint Receptor Lilrbs
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2019-11-27 Classification: IMMUNE SYSTEM Ligands: IOD |
 |
Organism: Protobothrops flavoviridis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-12-12 Classification: TOXIN/ANTITOXIN Ligands: MES, GOL |
 |
Crystal Structure Of Extracellular Domain Of Human Lectin-Like Transcript 1 (Llt1), The Ligand For Natural Killer Receptor-P1A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2015-06-24 Classification: IMMUNE SYSTEM Ligands: ZN, ACT, SO4 |
 |
Crystal Structure Of Hla-B*5201 In Complexed With Hiv Immunodominant Epitope (Taftipsi)
Organism: Homo sapiens, Human immunodeficiency virus type 1 (z2/cdc-z34 isolate)
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2013-02-13 Classification: IMMUNE SYSTEM |
 |
Organism: Chryseobacterium proteolyticum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-08-11 Classification: HYDROLASE |
 |
Organism: Chryseobacterium proteolyticum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-08-11 Classification: HYDROLASE |
 |
Organism: Chryseobacterium proteolyticum
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2010-08-11 Classification: HYDROLASE Ligands: CIT |
 |
Organism: Chryseobacterium proteolyticum
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2009-03-17 Classification: HYDROLASE |
 |
Organism: 'arthromyces ramosus'
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1997-03-12 Classification: OXIDOREDUCTASE Ligands: CA, HEM |
 |
Crystal Structures Of Cyanide-And Triiodide-Bound Forms Of Arthromyces Ramosus Peroxidase At Different Ph Values. Perturbations Of Active Site Residues And Their Implication In Enzyme Catalysis
Organism: 'arthromyces ramosus'
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 1996-01-29 Classification: PEROXIDASE (DONOR:H2O2 OXIDOREDUCTASE) Ligands: CYN, CA, HEM |
 |
Crystal Structures Of Cyanide-And Triiodide-Bound Forms Of Arthromyces Ramosus Peroxidase At Different Ph Values. Perturbations Of Active Site Residues And Their Implication In Enzyme Catalysis
Organism: 'arthromyces ramosus'
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 1996-01-29 Classification: PEROXIDASE (DONOR:H2O2 OXIDOREDUCTASE) Ligands: CYN, CA, HEM |
 |
Crystal Structures Of Cyanide-And Triiodide-Bound Forms Of Arthromyces Ramosus Peroxidase At Different Ph Values. Perturbations Of Active Site Residues And Their Implication In Enzyme Catalysis
Organism: 'arthromyces ramosus'
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 1996-01-29 Classification: PEROXIDASE (DONOR:H2O2 OXIDOREDUCTASE) Ligands: CYN, CA, HEM |
 |
Crystal Structures Of Cyanide-And Triiodide-Bound Forms Of Arthromyces Ramosus Peroxidase At Different Ph Values. Perturbations Of Active Site Residues And Their Implication In Enzyme Catalysis
Organism: 'arthromyces ramosus'
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1996-01-29 Classification: PEROXIDASE (DONOR:H2O2 OXIDOREDUCTASE) Ligands: CA, IOD, K, HEM |
 |
Crystal Structures Of Cyanide-And Triiodide-Bound Forms Of Arthromyces Ramosus Peroxidase At Different Ph Values. Perturbations Of Active Site Residues And Their Implication In Enzyme Catalysis
Organism: 'arthromyces ramosus'
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1996-01-29 Classification: PEROXIDASE (DONOR:H2O2 OXIDOREDUCTASE) Ligands: CA, IOD, K, HEM |
 |
Tertiary Structure Of [2Fe-2S] Ferredoxin From Spirulina Platensis Refined At 2.5 Angstroms Resolution: Structural Comparisons Of Plant-Type Ferredoxins And An Electrostatic Potential Analysis
Organism: Arthrospira platensis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1995-12-07 Classification: ELECTRON TRANSPORT Ligands: FES |
 |
Crystal Structure Of The Fungal Peroxidase From Arthromyces Ramosus At 1.9 Angstroms Resolution: Structural Comparisons With The Lignin And Cytochrome C Peroxidases
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1994-01-31 Classification: PEROXIDASE(DONOR:H2O2 OXIDOREDUCTASE) Ligands: CA, HEM |

