Search Count: 87
 |
Crystal Structure Of Manganese-Rebound N(Omega)-Hydroxy-L-Arginine Hydrolase With Oxidized Cys86
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2024-11-20 Classification: HYDROLASE Ligands: MN, MG |
 |
Crystal Structure Of Copper-Bound N(Omega)-Hydroxy-L-Arginine Hydrolase With Oxidized Cys86
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-11-20 Classification: HYDROLASE Ligands: MN, MG, CU, CU1 |
 |
Crystal Structure Of Copper-Bound N(Omega)-Hydroxy-L-Arginine Hydrolase Without Oxidized Cys86
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2024-11-20 Classification: HYDROLASE Ligands: MN, MG, CU1 |
 |
Crystal Structure Of Manganese-Free N(Omega)-Hydroxy-L-Arginine Hydrolase With Oxidized Cys86.
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-11-20 Classification: HYDROLASE Ligands: MN, MG |
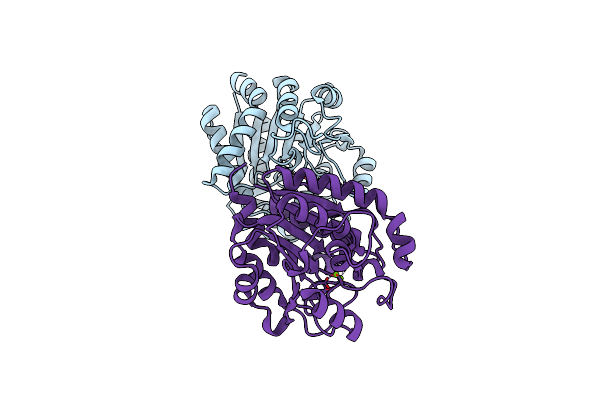 |
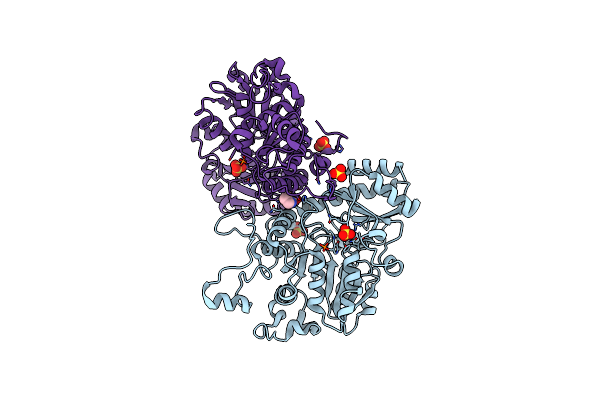
Crystal Structure Of Manganese-Free N(Omega)-Hydroxy-L-Arginine Hydrolase Without Oxidized Cys86
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2024-11-20 Classification: HYDROLASE Ligands: MN, MG |
 |
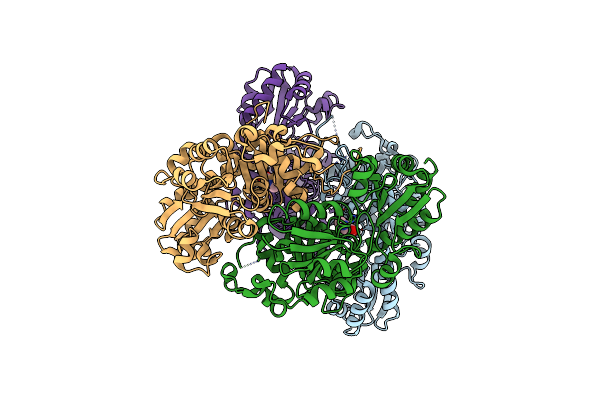
Crystal Structure Of O-Acetylhomoserine Sulfhydrylase From Lactobacillus Plantarum In The Closed Form
Organism: Lactiplantibacillus plantarum jdm1
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2024-02-14 Classification: LYASE Ligands: SO4, PRO |
 |
Crystal Structure Of O-Acetylhomoserine Sulfhydrylase From Lactobacillus Plantarum In The Open Form
Organism: Lactiplantibacillus plantarum jdm1
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-02-14 Classification: LYASE Ligands: PRO |
 |
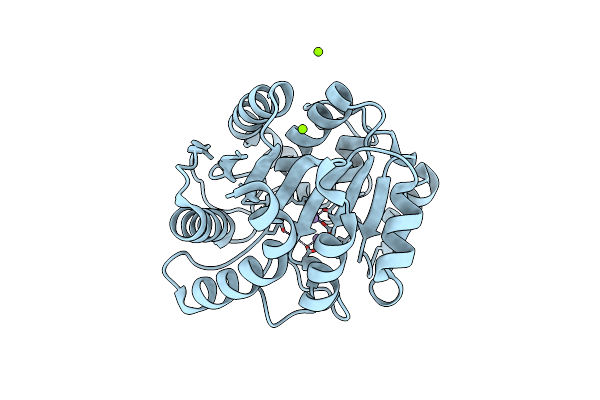
Crystal Structure Of N(Omega)-Hydroxy-L-Arginine Hydrolase In Complex With L-Orn
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-05-18 Classification: ANTIBIOTIC Ligands: MN, MG, ORN |
 |
Crystal Structure Of C86H-H196S Mutant Of N(Omega)-Hydroxy-L-Arginine Hydrolase
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2022-05-18 Classification: ANTIBIOTIC Ligands: MN, MG |
 |
Crystal Structure Of N(Omega)-Hydroxy-L-Arginine Hydrolase In Complex With Abh
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2022-05-18 Classification: ANTIBIOTIC Ligands: MN, MG, ABH |
 |
Crystal Structure Of C86H-Y124N-G126H-H196S Mutant Of N(Omega)-Hydroxy-L-Arginine Hydrolase
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-05-18 Classification: ANTIBIOTIC Ligands: MN, MG |
 |
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2021-12-15 Classification: ISOMERASE Ligands: SO4 |
 |
Crystal Structure Of Tyrosinase From Streptomyces Castaneoglobisporus In Complex With The Caddie Protein Obtained By Soaking In The Solution Containing Cu(Ii) And Hydroxylamine For 24 H
Organism: Streptomyces castaneoglobisporus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-06-16 Classification: METAL BINDING PROTEIN Ligands: PEO, CU, NO3 |
 |
Crystal Structure Of N191G-Mutated Tyrosinase From Streptomyces Castaneoglobisporus In Complex With The Caddie Protein Obtained By Soaking In The Solution Containing Cu(Ii) And Hydroxylamine For 24 H
Organism: Streptomyces castaneoglobisporus
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2021-06-16 Classification: METAL BINDING PROTEIN Ligands: PEO, CU, NO3 |
 |
Organism: Pseudomonas taiwanensis dsm 21245
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2021-04-28 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
 |
Organism: Pseudomonas taiwanensis dsm 21245
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 2021-04-28 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
 |
Crystal Structure Of Cystathionine Gamma-Lyase From Lactobacillus Plantarum Complexed With L-Serine
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2020-10-07 Classification: LYASE Ligands: KOU, PO4 |
 |
Crystal Structure Of Cystathionine Gamma-Lyase From Lactobacillus Plantarum Complexed With Cystathionine
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2020-10-07 Classification: LYASE Ligands: E9U, PO4 |
 |
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-09-02 Classification: HYDROLASE Ligands: MN, MG |
 |
Organism: Homo sapiens, Murine respirovirus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-08-19 Classification: VIRAL PROTEIN |

