Search Count: 130
 |
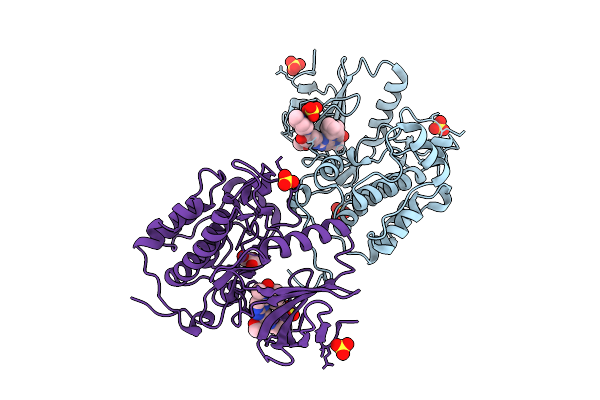
Plk1 Surface Entropy Reduction (Ser) Mutant In Complex With Thiazolidinone Inhibitor Compound 1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: A1JCP, SO4, GOL, CL |
 |
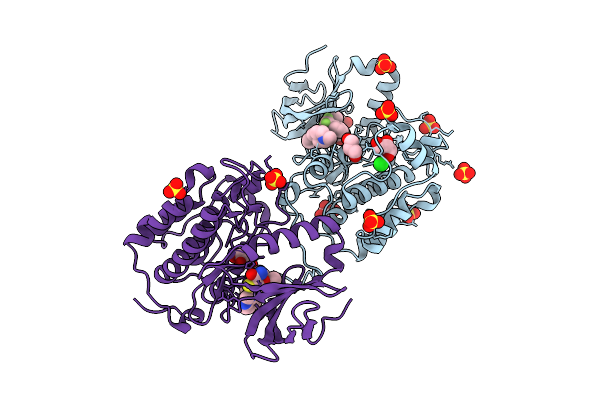
Plk1 Surface Entropy Reduction (Ser) Mutant In Complex With Inhibitor Bi 2536
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: R78, SO4, GOL |
 |
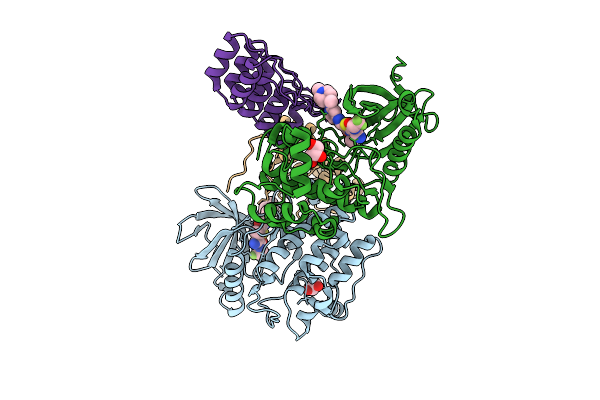
Plk1 Surface Entropy Reduction (Ser) Mutant In Complex With Inhibitor Gsk461364
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: A1JCQ, SO4, CL, GOL |
 |
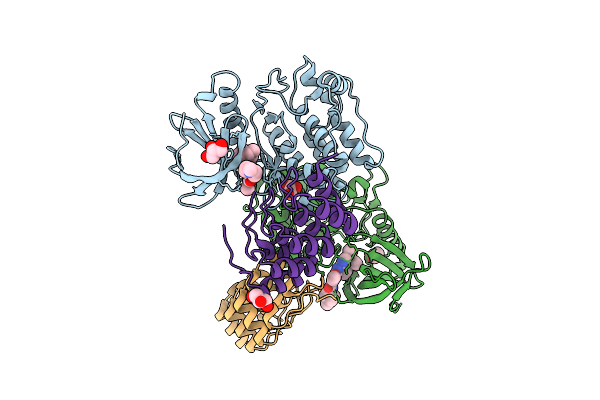
Crystal Structure Of Wild-Type Plk1 Kinase Domain In Complex With Thiazolidinone Inhibitor Compound 1 And A Selective Darpin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: A1JCP, GOL, CL |
 |
Crystal Structure Of Wild-Type Plk1 Kinase Domain In Complex With Thiazolidinone Inhibitor Compound 1 And A Selective Darpin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: R78, GOL |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: METAL BINDING PROTEIN Ligands: FE, O, ZN |
 |
Organism: Clostridioides difficile p28
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: OXIDOREDUCTASE Ligands: FE, O |
 |
Ferric-Siderophore Reduction In Shewanella Biscestrii: Structural And Functional Characterization Of Sbisip Reveals Unforeseen Specificity
Organism: Shewanella bicestrii
Method: X-RAY DIFFRACTION Release Date: 2024-01-17 Classification: METAL BINDING PROTEIN Ligands: FAD |
 |
Structure Of The C-Terminally Truncated Nad+-Dependent Dna Ligase From The Poly-Extremophile Deinococcus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:3.36 Å Release Date: 2023-09-27 Classification: DNA BINDING PROTEIN Ligands: MN, ZN |
 |
Arsenate Reductase (Arsc2) From Deinococcus Indicus, Co-Crystallized With Arsenate
Organism: Deinococcus indicus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-08-30 Classification: OXIDOREDUCTASE Ligands: ART, GOL |
 |
Organism: Deinococcus indicus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-08-30 Classification: OXIDOREDUCTASE |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2023-07-12 Classification: OXIDOREDUCTASE Ligands: FES |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2022-07-06 Classification: METAL BINDING PROTEIN Ligands: MN, O, GOL, SO4, CL |
 |
Structure Of Bg10: An Alcohol-Tolerant And Glucose-Stimulated B-Glucosidase
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-01-12 Classification: HYDROLASE |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2022-01-12 Classification: METAL BINDING PROTEIN Ligands: MN, O, SO4, GOL |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2021-04-21 Classification: METAL BINDING PROTEIN Ligands: FE, O, GOL |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2021-04-21 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2021-04-21 Classification: METAL BINDING PROTEIN Ligands: FE, O, GOL |
 |
Structure Of [Nifese] Hydrogenase From Desulfovibrio Vulgaris Hildenborough Pressurized With Krypton Gas - Structure Wtkr1
Organism: Desulfovibrio vulgaris (strain hildenborough / atcc 29579 / dsm 644 / ncimb 8303)
Method: X-RAY DIFFRACTION Release Date: 2020-09-09 Classification: OXIDOREDUCTASE Ligands: SF4, 6ML, KR, FCO, NI, FE2, H2S, CL |
 |
Structure Of [Nifese] Hydrogenase From Desulfovibrio Vulgaris Hildenborough Pressurized With Oxygen Gas - Structure Wto2
Organism: Desulfovibrio vulgaris (strain hildenborough / atcc 29579 / dsm 644 / ncimb 8303)
Method: X-RAY DIFFRACTION Release Date: 2020-09-09 Classification: OXIDOREDUCTASE Ligands: SF4, 6ML, GOL, FCO, NI, FE2, H2S, CL, OXY |

