Search Count: 23
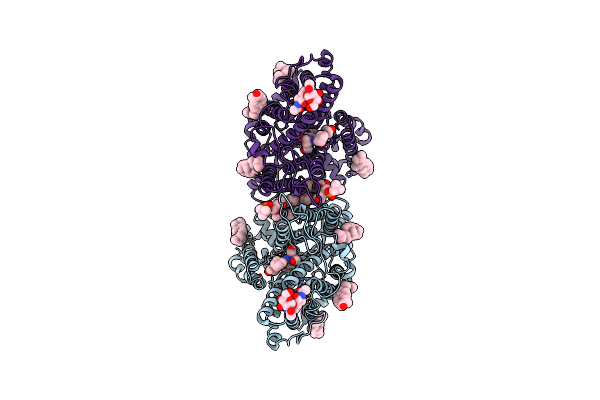 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.13 Å Release Date: 2023-09-13 Classification: TRANSPORT PROTEIN Ligands: CLR, PC1, H9F |
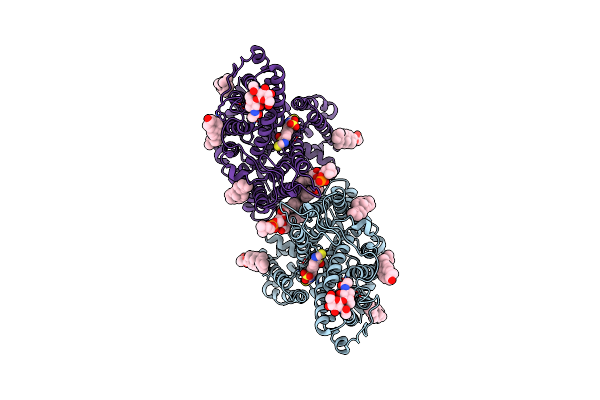 |
Cryo-Em Structure Of Human Anion Exchanger 1 Bound To 4,4'-Diisothiocyanatostilbene-2,2'-Disulfonic Acid (Dids)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.95 Å Release Date: 2023-09-13 Classification: TRANSPORT PROTEIN Ligands: 4DS, CLR, Y01, PC1 |
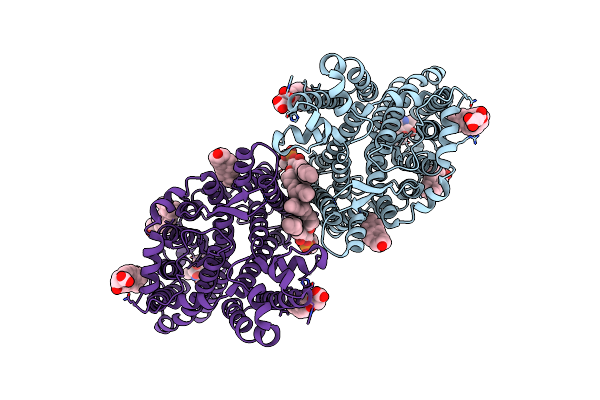 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.99 Å Release Date: 2023-08-16 Classification: TRANSPORT PROTEIN Ligands: CLR, PC1, Y01 |
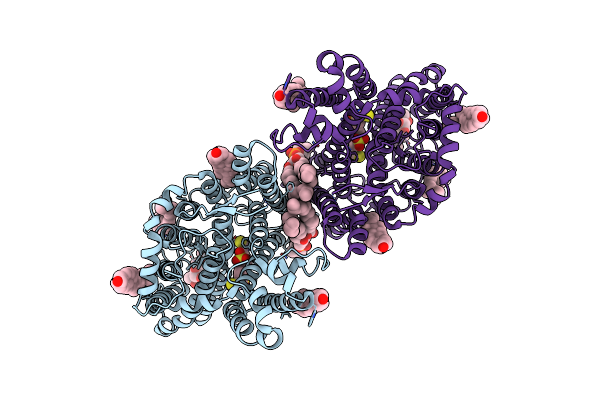 |
Cryo-Em Structure Of Human Anion Exchanger 1 Bound To 4,4'-Diisothiocyanatodihydrostilbene-2,2'-Disulfonic Acid (H2Dids)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.98 Å Release Date: 2023-08-16 Classification: TRANSPORT PROTEIN Ligands: CLR, PC1, 4DS |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.37 Å Release Date: 2023-08-16 Classification: TRANSPORT PROTEIN Ligands: BCT, CLR, PC1 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.18 Å Release Date: 2023-08-16 Classification: TRANSPORT PROTEIN Ligands: CLR, PC1, NFL |
 |
Cryo-Em Structure Of Human Anion Exchanger 1 Modified With Diethyl Pyrocarbonate (Depc)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.07 Å Release Date: 2023-08-16 Classification: TRANSPORT PROTEIN Ligands: CLR, PC1 |
 |
Cryoem Structure Of Girk2Pip2* - G Protein-Gated Inwardly Rectifying Potassium Channel Girk2 With Pip2
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2021-09-01 Classification: TRANSPORT PROTEIN Ligands: PIO, K, NA |
 |
Cryoem Structure Of Girk2-Pip2/Chs - G Protein-Gated Inwardly Rectifying Potassium Channel Girk2 With Modulators Cholesteryl Hemisuccinate And Pip2
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2021-09-01 Classification: TRANSPORT PROTEIN Ligands: PIO, Y01, K, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2021-02-24 Classification: SIGNALING PROTEIN Ligands: GOL, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2021-02-24 Classification: SIGNALING PROTEIN Ligands: NAG |
 |
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2018-04-25 Classification: VIRAL PROTEIN/RNA |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2017-06-07 Classification: OXIDOREDUCTASE Ligands: ZN, GOL |
 |
Crystal Structure Of F222 Form Of E112A Mutant Of Stationary Phase Survival Protein (Sure) From Salmonella Typhimurium
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2015-09-09 Classification: HYDROLASE Ligands: MG, PO4, EDO |
 |
Crystal Structure Of C2 Form Of E112A Mutant Of Stationary Phase Survival Protein (Sure) From Salmonella Typhimurium
Organism: Salmonella enterica subsp. enterica serovar enteritidis
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2015-09-09 Classification: HYDROLASE Ligands: PO4, MG, GOL, MPD |
 |
Crystal Structure Of F222 Form Of E112A/H234A Mutant Of Stationary Phase Survival Protein (Sure) From Salmonella Typhimurium
Organism: Salmonella typhimurium lt2
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-09-09 Classification: HYDROLASE Ligands: MG, PO4, EDO |
 |
Crystal Structure Of C2 Form Of E112A/H234A Mutant Of Stationary Phase Survival Protein (Sure) From Salmonella Typhimurium
Organism: Salmonella typhimurium lt2
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2015-09-09 Classification: HYDROLASE Ligands: ACT, CA, PO4, MPD, MG |
 |
Crystal Structure Of E112A/H234A Mutant Of Stationary Phase Survival Protein (Sure) From Salmonella Typhimurium Co-Crystallized With Amp
Organism: Salmonella typhimurium lt2
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2015-09-09 Classification: HYDROLASE Ligands: MG, PO4, MPD |
 |
Crystal Structure Of E112A/H234A Mutant Of Stationary Phase Survival Protein (Sure) From Salmonella Typhimurium Co-Crystallized And Soaked With Amp.
Organism: Salmonella typhimurium lt2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-09-09 Classification: HYDROLASE Ligands: MG, PO4, ADE, EDO |
 |
Crystal Structure Of E112A/D230A Mutant Of Stationary Phase Survival Protein (Sure) From Salmonella Typhimurium
Organism: Salmonella typhimurium lt2
Method: X-RAY DIFFRACTION Resolution:3.56 Å Release Date: 2015-09-09 Classification: HYDROLASE |

