Search Count: 123
 |
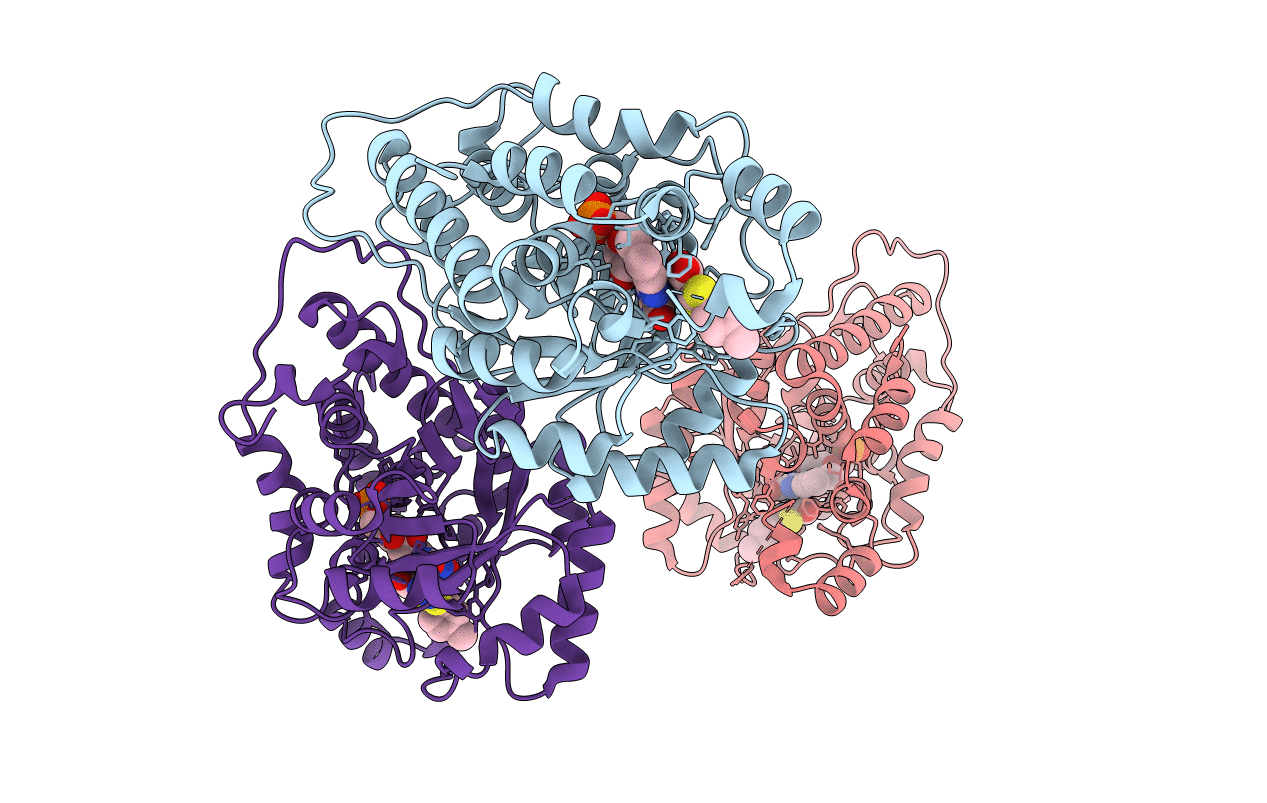
Crystal Structure Of Monotopic Membrane Protein (S)-Mandelate Dehydrogenase
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-08-15 Classification: OXIDOREDUCTASE Ligands: FMN, GOL, SO4, EDO, CIT, PEG, PO4, NA, LMT |
 |
High Resolution Crystal Structure Of Rat Long Chain Hydroxy Acid Oxidase In Complex With The Inhibitor 4-Carboxy-5-[(4-Chiorophenyl)Sulfanyl]-1, 2, 3-Thiadiazole.
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2012-03-07 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: FMN, HO6 |
 |
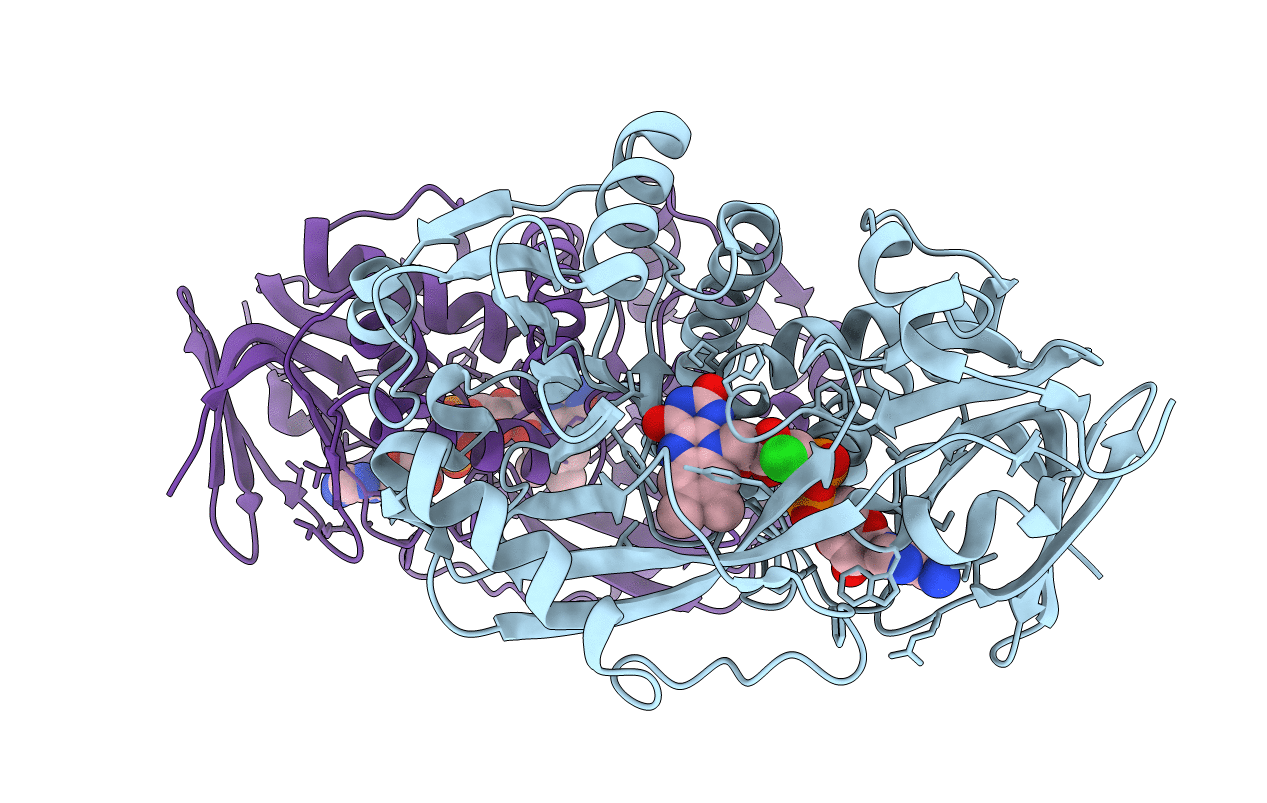
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2011-06-01 Classification: OXIDOREDUCTASE Ligands: FAD, CL, SAR |
 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-06-01 Classification: OXIDOREDUCTASE Ligands: FAD, CL |
 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2011-06-01 Classification: OXIDOREDUCTASE Ligands: FAD, CL, MTG |
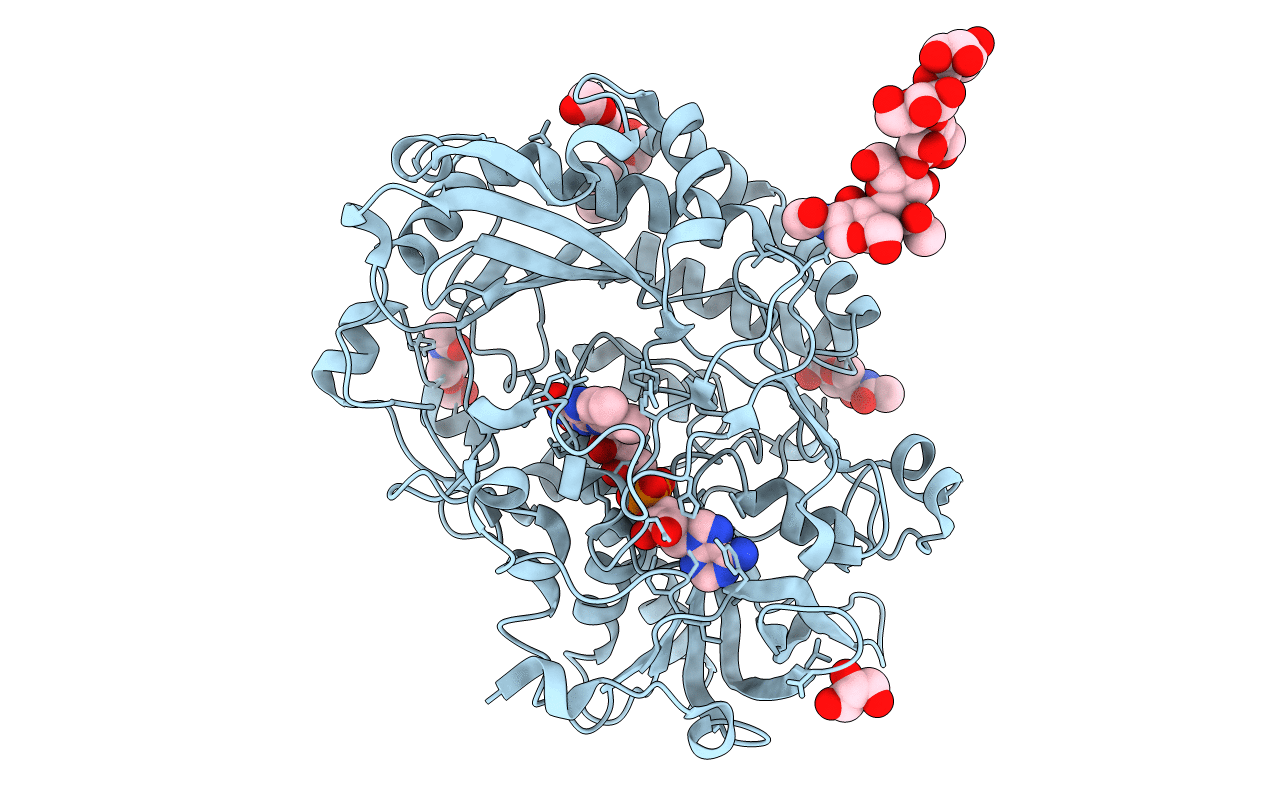 |
Crystal Structure Of Glucose Oxidase For Space Group C2221 At 1.2 A Resolution
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2011-06-01 Classification: OXIDOREDUCTASE Ligands: NAG, FAD, GOL, PEG, CL |
 |
Crystal Structure Of Glucose Oxidase For Space Group P3121 At 1.3 A Resolution.
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2011-06-01 Classification: OXIDOREDUCTASE Ligands: NAG, FAD, CL |
 |
Organism: Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2011-02-09 Classification: ELECTRON TRANSPORT Ligands: CU, NA, PO4, K |
 |
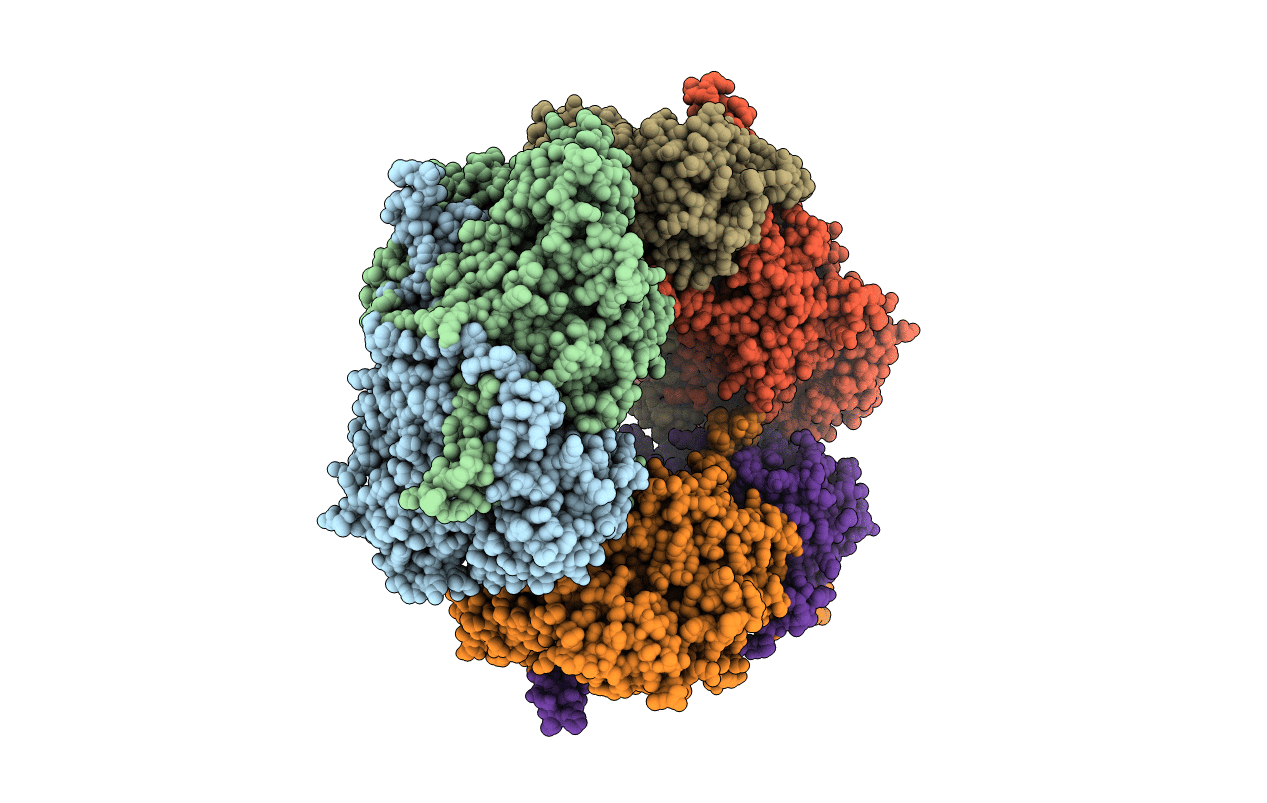
Crystal Structural Of Mutant Y305A In The Copper Amine Oxidase From Hansenula Polymorpha
Organism: Pichia angusta
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-08-25 Classification: OXIDOREDUCTASE Ligands: CU |
 |
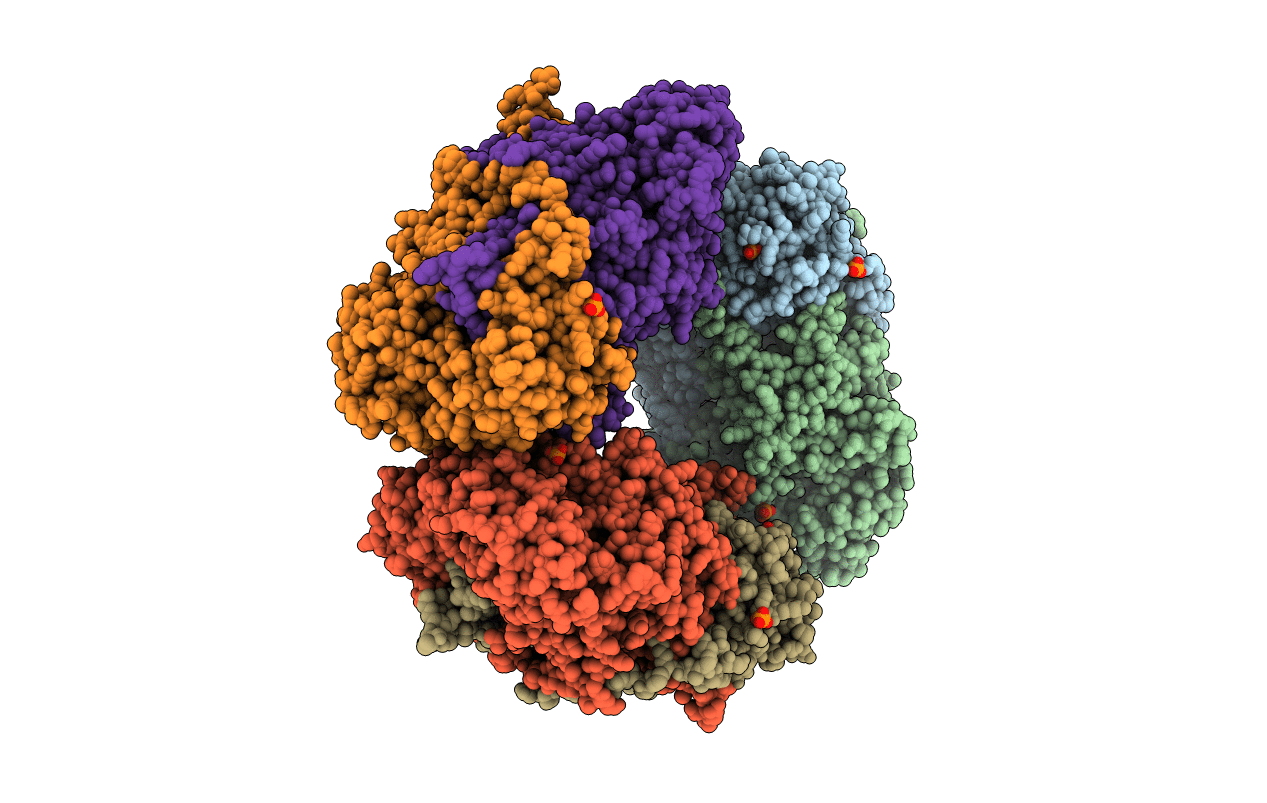
Crystal Structure Of Mutant Y305F Expressed In E. Coli In The Copper Amine Oxidase From Hansenula Polymorpha
Organism: Pichia angusta
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2010-08-25 Classification: OXIDOREDUCTASE Ligands: CU |
 |
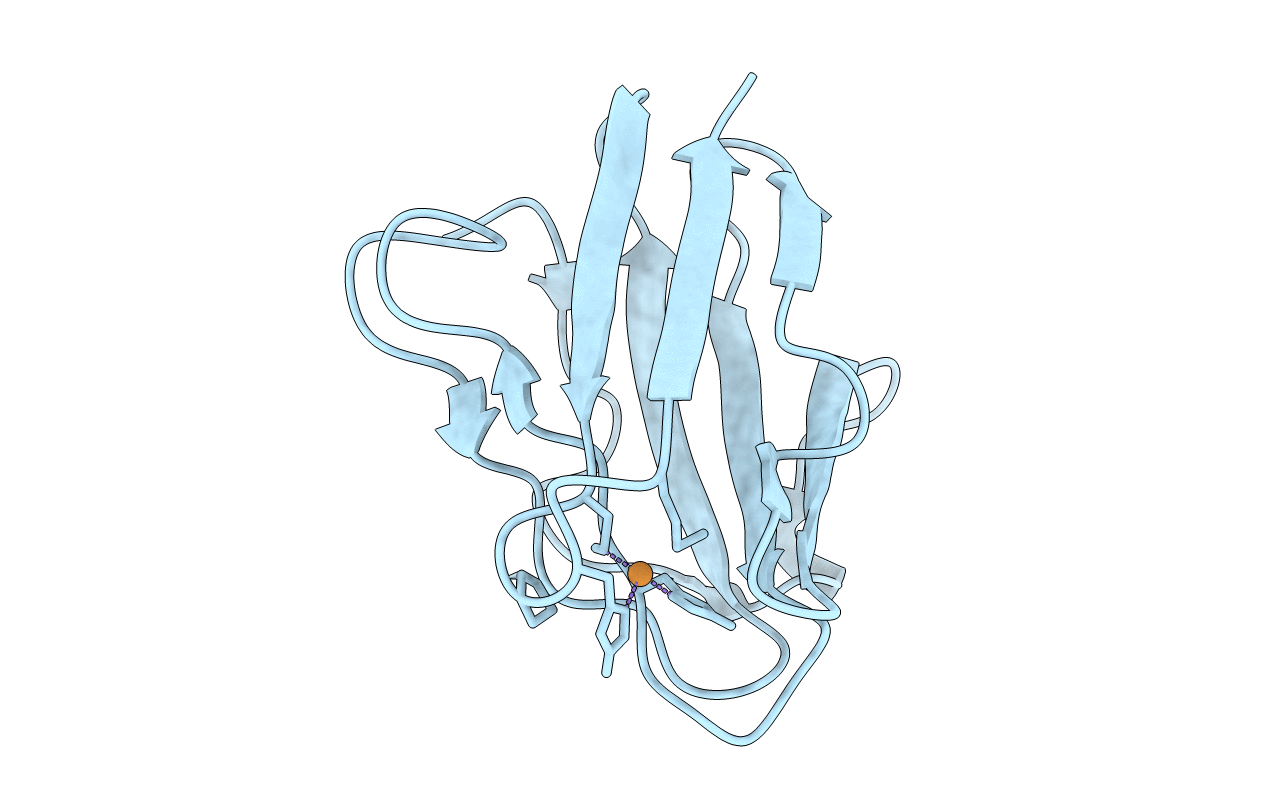
Crystal Structure Of Y305F Mutant Of The Copper Amine Oxidase From Hansenula Polymorpha Expressed In Yeast
Organism: Pichia angusta
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-08-25 Classification: OXIDOREDUCTASE Ligands: CU, PO4 |
 |
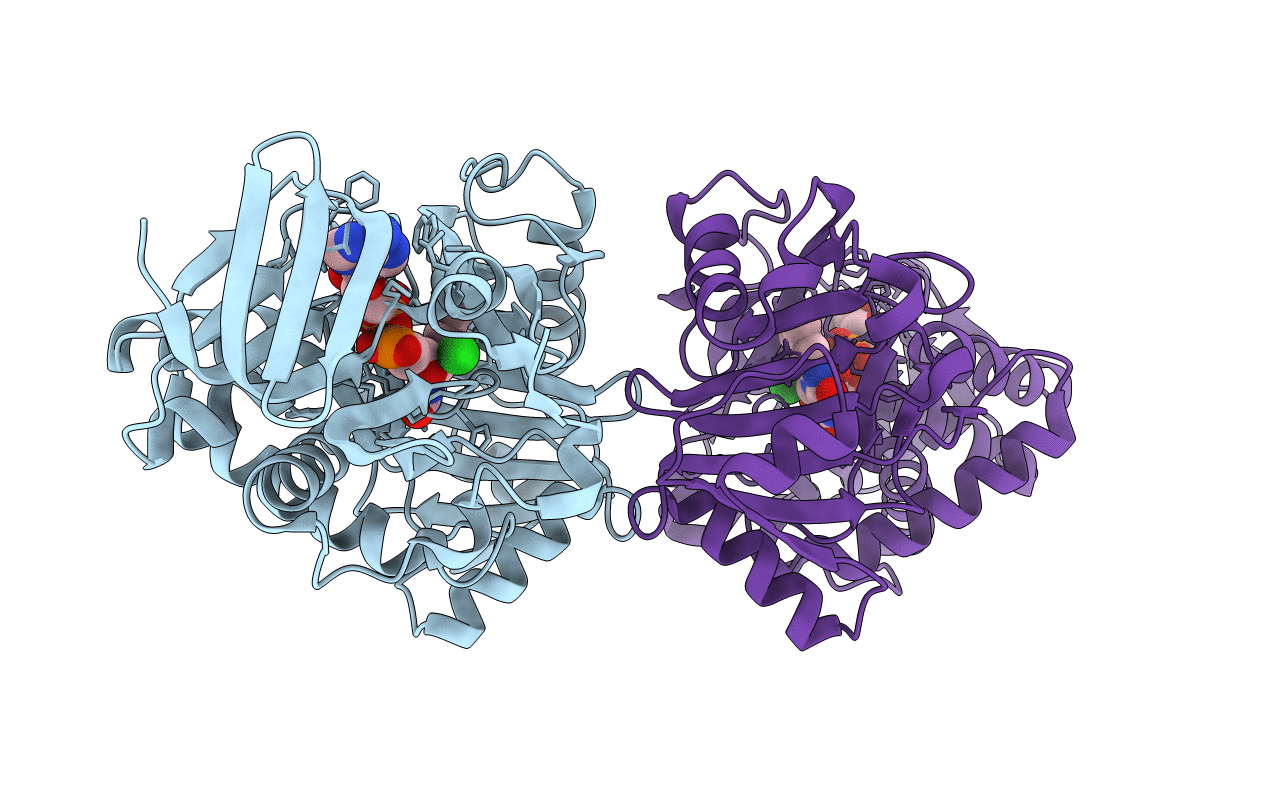
Organism: Paracoccus denitrificans
Method: NEUTRON DIFFRACTION, X-RAY DIFFRACTION Resolution:1.80 Å, 1.5 Å Release Date: 2010-04-28 Classification: ELECTRON TRANSPORT Ligands: CU, DOD |
 |
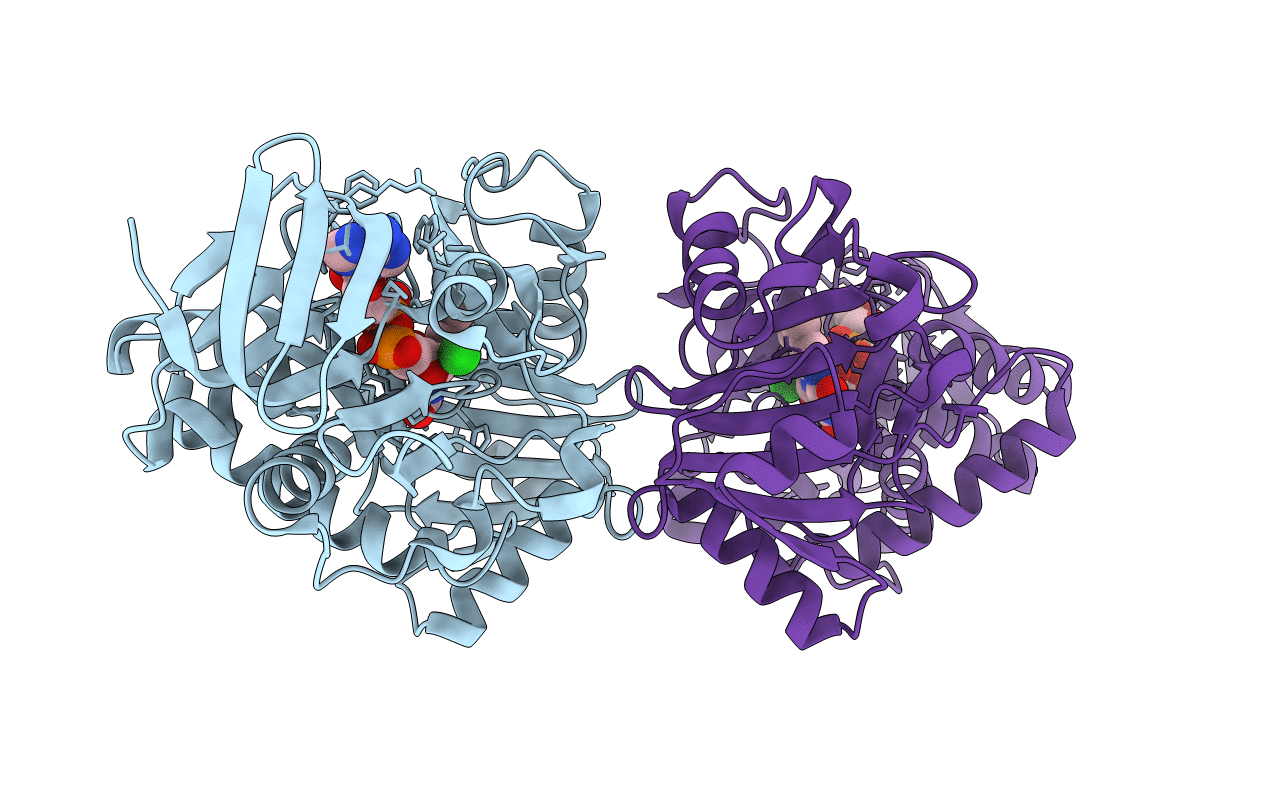
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: FAD, CL |
 |
Crystal Structure Of The Lys265Arg Phosphate-Crytsallized Mutant Of Monomeric Sarcosine Oxidase
Organism: Bacillus sp. b-0618
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: FAD, CL |
 |
Crystal Structure Of The Lys265Arg Peg-Crystallized Mutant Of Monomeric Sarcosine Oxidase
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: FAD, CL, PO4 |
 |
Crystal Structures Of The G81A Mutant Of The Active Chimera Of (S)-Mandelate Dehydrogenase And Its Complex With Two Of Its Substrates
Organism: Pseudomonas putida, Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2009-12-22 Classification: OXIDOREDUCTASE Ligands: FMN, MES |
 |
Tyr258Phe Mutant Of Nikd, An Unusual Amino Acid Oxidase Essential For Nikkomycin Biosynthesis: Open Form At 1.55A Resolution
Organism: Streptomyces tendae
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2009-10-20 Classification: OXIDOREDUCTASE Ligands: FAD, 6PC, MPD |
 |
Crystal Structure Of R49K Mutant Of Monomeric Sarcosine Oxidase Crystallized In Peg As Precipitant
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-02-26 Classification: OXIDOREDUCTASE Ligands: CL, FAD |
 |
Crystal Structure Of R49K Mutant Of Monomeric Sarcosine Oxidase Crystallized In Phosphate As Precipitant
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2008-02-26 Classification: OXIDOREDUCTASE Ligands: CL, FAD, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-01-01 Classification: HYDROLASE Ligands: NAG |

